








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (9): 124-138.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0103
LIU Jia-li( ), SONG Jing-rong, ZHAO Wen-yu, ZHANG Xin-yuan, ZHAO Zi-yang, CAO Yi-bo(
), SONG Jing-rong, ZHAO Wen-yu, ZHANG Xin-yuan, ZHAO Zi-yang, CAO Yi-bo( ), ZHANG Ling-yun(
), ZHANG Ling-yun( )
)
Received:2025-01-23
Online:2025-09-26
Published:2025-09-24
Contact:
CAO Yi-bo, ZHANG Ling-yun
E-mail:936606393@qq.com;caoyibo@bjfu.edu.cn;lyzhang@bjfu.edu.cn
LIU Jia-li, SONG Jing-rong, ZHAO Wen-yu, ZHANG Xin-yuan, ZHAO Zi-yang, CAO Yi-bo, ZHANG Ling-yun. Identification of the R2R3-MYB Gene and Expression Analysis of Flavonoid Regulatory Genes in Blueberry[J]. Biotechnology Bulletin, 2025, 41(9): 124-138.
基因 Gene | 引物 Primer (5'-3') | ∆G/∆g (25 ℃) (kcal/mol) | ∆H/∆h (kcal/mol) | 3'∆G (kcal/mol) | Tm (℃) | GC (%) | Primer score | PCR score |
|---|---|---|---|---|---|---|---|---|
| VcMYB44 | F: GACCAGACCTAAAACGAGGCAAC | -36.5/-35.0 | -190.5/-181.6 | -7.3 | 60.8 | 52.2 | 941 | 920 |
| R: GGCAATCTTCCTGCTATTAGTGACC | -37.9/-36.7 | -201.5/-194.0 | -7.1 | 60.4 | 48.0 | 963 | ||
| VcMYB56 | F: TCGGAATATCCTAACGACCAACCAC | -37.9/-37.2 | -202.2/-194.5 | -7.2 | 60.3 | 48.0 | 946 | 889 |
| R: GACTTTCTGCTTTCGGGCCTC | -34.0/-32.9 | -173.0/-166.5 | -7.7 | 60.8 | 57.1 | 856 | ||
| VcMYB77 | F: TTCTTCCGATCAGGACAAACCCTC | -36.9/-36.0 | -193.2/-185.4 | -7.3 | 60.9 | 50.0 | 940 | 877 |
| R: TGACACTGCCGTGGTAATACCC | -34.5/-34.0 | -179.3/-171.5 | -6.7 | 60.0 | 54.5 | 897 | ||
| VcMYB88 | F: TTGCCTTTCCCTCTCCTCTAATGC | -36.6/-35.9 | -191.7/-184.0 | -6.7 | 60.6 | 50.0 | 880 | 900 |
| R: TAGGAATCCCAAGACTTTCCGTTT | -34.5/-34.3 | -177.7/-181.3 | -6.8 | 60.5 | 41.7 | 920 | ||
| VcMYB91 | F: CCACCTGACCGACAAATGCAA | -33.3/-32.7 | -168.9/-164.8 | -7.3 | 60.6 | 52.4 | 930 | 926 |
| R: AACCGTGCATATCCCTTACGAC | -34.5/-33.2 | -177.3/-172.0 | -7.5 | 60.5 | 50.0 | 941 | ||
| VcMYB96 | F: CCTTATCAAAACCAATCTGAACCGT | -37.0/-35.6 | -194.0/-192.1 | -8.0 | 60.9 | 40.0 | 839 | 867 |
| R: TTTGCTACTGCCACTATTCCCAC | -35.1/-34.2 | -182.7/-177.3 | -7.6 | 60.3 | 47.8 | 901 | ||
| VcMYB103 | F: TCACGCGCTGAATCCTAACCC | -34.2/-33.5 | -174.1/-166.6 | -7.1 | 61.0 | 57.1 | 936 | 883 |
| R: AACACTGCCGTGGTAATACCC | -32.9/-32.0 | -167.4/-162.7 | -6.7 | 60.1 | 52.4 | 889 | ||
| VcMYB139 | F: GTTGAGAGATCCCTCCGACCA | -32.9/-32.1 | -167.1/-161.7 | -7.1 | 60.3 | 57.1 | 920 | 888 |
| R: CTTTTCGGTCAGTAGGACTTCGTT | -36.0/-35.4 | -187.2/-187.6 | -7.1 | 60.8 | 45.8 | 933 | ||
| VcMYB172 | F: CCTCGCTCTCAAAATCCACGTC | -35.4/-34.2 | -183.7/-175.9 | -7.5 | 60.5 | 54.5 | 928 | 879 |
| R: TCTAGACAACTTGCAGCATCGG | -33.7/-33.4 | -171.9/-172.6 | -7.3 | 60.5 | 50.0 | 954 | ||
| VcMYB214 | F: GAAAGTCCATAACCCAAAGCCCAA | -36.1/-35.4 | -188.9/-183.5 | -7.1 | 60.6 | 45.8 | 918 | 893 |
| R: TCTTTAGGAGGGCAATCAAAGCTAC | -37.0/-36.1 | -195.5/-191.3 | -6.6 | 60.3 | 44.0 | 867 | ||
| VcMYB229 | F: TGAATTACCTCCGTCCGAACGTC | -36.3/-35.5 | -191.1/-182.4 | -7.5 | 60.3 | 52.2 | 920 | 793 |
| R: TCTCCCAGCTATCAGCGACCA | -32.8/-32.9 | -166.6/-161.9 | -7.1 | 60.2 | 57.1 | 919 | ||
| VcMYB236 | F: AGTTGTAATGCCGTCAACACCGA | -35.7/-35.5 | -186.5/-180.7 | -7.9 | 60.5 | 47.8 | 887 | 857 |
| R: ACTAGTGCCGTGATCATTTATTCCG | -38.1/-36.7 | -201.0/-195.2 | -7.4 | 61.0 | 44.0 | 904 | ||
| VcMYB256 | F: ACTACTGGAACACCCATATCCGAA | -35.4/-35.0 | -183.1/-181.9 | -7.4 | 60.8 | 45.8 | 937 | 861 |
| R: ATTAGCAGTCGCAAAAGATATGGT | -34.7/-34.0 | -180.2/-183.3 | -6.6 | 60.1 | 37.5 | 879 | ||
| VcMYB259 | F: TCCTTAATTGCGGGTAGACTTCCA | -35.1/-35.3 | -182.2/-182.8 | -6.7 | 60.3 | 45.8 | 887 | 866 |
| R: TCCTTTCAGTCGTATTCAAGGTCG | -36.1/-35.3 | -189.0/-187.4 | -7.9 | 60.4 | 45.8 | 866 |
Table 1 Primers used for RT-qPCR
基因 Gene | 引物 Primer (5'-3') | ∆G/∆g (25 ℃) (kcal/mol) | ∆H/∆h (kcal/mol) | 3'∆G (kcal/mol) | Tm (℃) | GC (%) | Primer score | PCR score |
|---|---|---|---|---|---|---|---|---|
| VcMYB44 | F: GACCAGACCTAAAACGAGGCAAC | -36.5/-35.0 | -190.5/-181.6 | -7.3 | 60.8 | 52.2 | 941 | 920 |
| R: GGCAATCTTCCTGCTATTAGTGACC | -37.9/-36.7 | -201.5/-194.0 | -7.1 | 60.4 | 48.0 | 963 | ||
| VcMYB56 | F: TCGGAATATCCTAACGACCAACCAC | -37.9/-37.2 | -202.2/-194.5 | -7.2 | 60.3 | 48.0 | 946 | 889 |
| R: GACTTTCTGCTTTCGGGCCTC | -34.0/-32.9 | -173.0/-166.5 | -7.7 | 60.8 | 57.1 | 856 | ||
| VcMYB77 | F: TTCTTCCGATCAGGACAAACCCTC | -36.9/-36.0 | -193.2/-185.4 | -7.3 | 60.9 | 50.0 | 940 | 877 |
| R: TGACACTGCCGTGGTAATACCC | -34.5/-34.0 | -179.3/-171.5 | -6.7 | 60.0 | 54.5 | 897 | ||
| VcMYB88 | F: TTGCCTTTCCCTCTCCTCTAATGC | -36.6/-35.9 | -191.7/-184.0 | -6.7 | 60.6 | 50.0 | 880 | 900 |
| R: TAGGAATCCCAAGACTTTCCGTTT | -34.5/-34.3 | -177.7/-181.3 | -6.8 | 60.5 | 41.7 | 920 | ||
| VcMYB91 | F: CCACCTGACCGACAAATGCAA | -33.3/-32.7 | -168.9/-164.8 | -7.3 | 60.6 | 52.4 | 930 | 926 |
| R: AACCGTGCATATCCCTTACGAC | -34.5/-33.2 | -177.3/-172.0 | -7.5 | 60.5 | 50.0 | 941 | ||
| VcMYB96 | F: CCTTATCAAAACCAATCTGAACCGT | -37.0/-35.6 | -194.0/-192.1 | -8.0 | 60.9 | 40.0 | 839 | 867 |
| R: TTTGCTACTGCCACTATTCCCAC | -35.1/-34.2 | -182.7/-177.3 | -7.6 | 60.3 | 47.8 | 901 | ||
| VcMYB103 | F: TCACGCGCTGAATCCTAACCC | -34.2/-33.5 | -174.1/-166.6 | -7.1 | 61.0 | 57.1 | 936 | 883 |
| R: AACACTGCCGTGGTAATACCC | -32.9/-32.0 | -167.4/-162.7 | -6.7 | 60.1 | 52.4 | 889 | ||
| VcMYB139 | F: GTTGAGAGATCCCTCCGACCA | -32.9/-32.1 | -167.1/-161.7 | -7.1 | 60.3 | 57.1 | 920 | 888 |
| R: CTTTTCGGTCAGTAGGACTTCGTT | -36.0/-35.4 | -187.2/-187.6 | -7.1 | 60.8 | 45.8 | 933 | ||
| VcMYB172 | F: CCTCGCTCTCAAAATCCACGTC | -35.4/-34.2 | -183.7/-175.9 | -7.5 | 60.5 | 54.5 | 928 | 879 |
| R: TCTAGACAACTTGCAGCATCGG | -33.7/-33.4 | -171.9/-172.6 | -7.3 | 60.5 | 50.0 | 954 | ||
| VcMYB214 | F: GAAAGTCCATAACCCAAAGCCCAA | -36.1/-35.4 | -188.9/-183.5 | -7.1 | 60.6 | 45.8 | 918 | 893 |
| R: TCTTTAGGAGGGCAATCAAAGCTAC | -37.0/-36.1 | -195.5/-191.3 | -6.6 | 60.3 | 44.0 | 867 | ||
| VcMYB229 | F: TGAATTACCTCCGTCCGAACGTC | -36.3/-35.5 | -191.1/-182.4 | -7.5 | 60.3 | 52.2 | 920 | 793 |
| R: TCTCCCAGCTATCAGCGACCA | -32.8/-32.9 | -166.6/-161.9 | -7.1 | 60.2 | 57.1 | 919 | ||
| VcMYB236 | F: AGTTGTAATGCCGTCAACACCGA | -35.7/-35.5 | -186.5/-180.7 | -7.9 | 60.5 | 47.8 | 887 | 857 |
| R: ACTAGTGCCGTGATCATTTATTCCG | -38.1/-36.7 | -201.0/-195.2 | -7.4 | 61.0 | 44.0 | 904 | ||
| VcMYB256 | F: ACTACTGGAACACCCATATCCGAA | -35.4/-35.0 | -183.1/-181.9 | -7.4 | 60.8 | 45.8 | 937 | 861 |
| R: ATTAGCAGTCGCAAAAGATATGGT | -34.7/-34.0 | -180.2/-183.3 | -6.6 | 60.1 | 37.5 | 879 | ||
| VcMYB259 | F: TCCTTAATTGCGGGTAGACTTCCA | -35.1/-35.3 | -182.2/-182.8 | -6.7 | 60.3 | 45.8 | 887 | 866 |
| R: TCCTTTCAGTCGTATTCAAGGTCG | -36.1/-35.3 | -189.0/-187.4 | -7.9 | 60.4 | 45.8 | 866 |
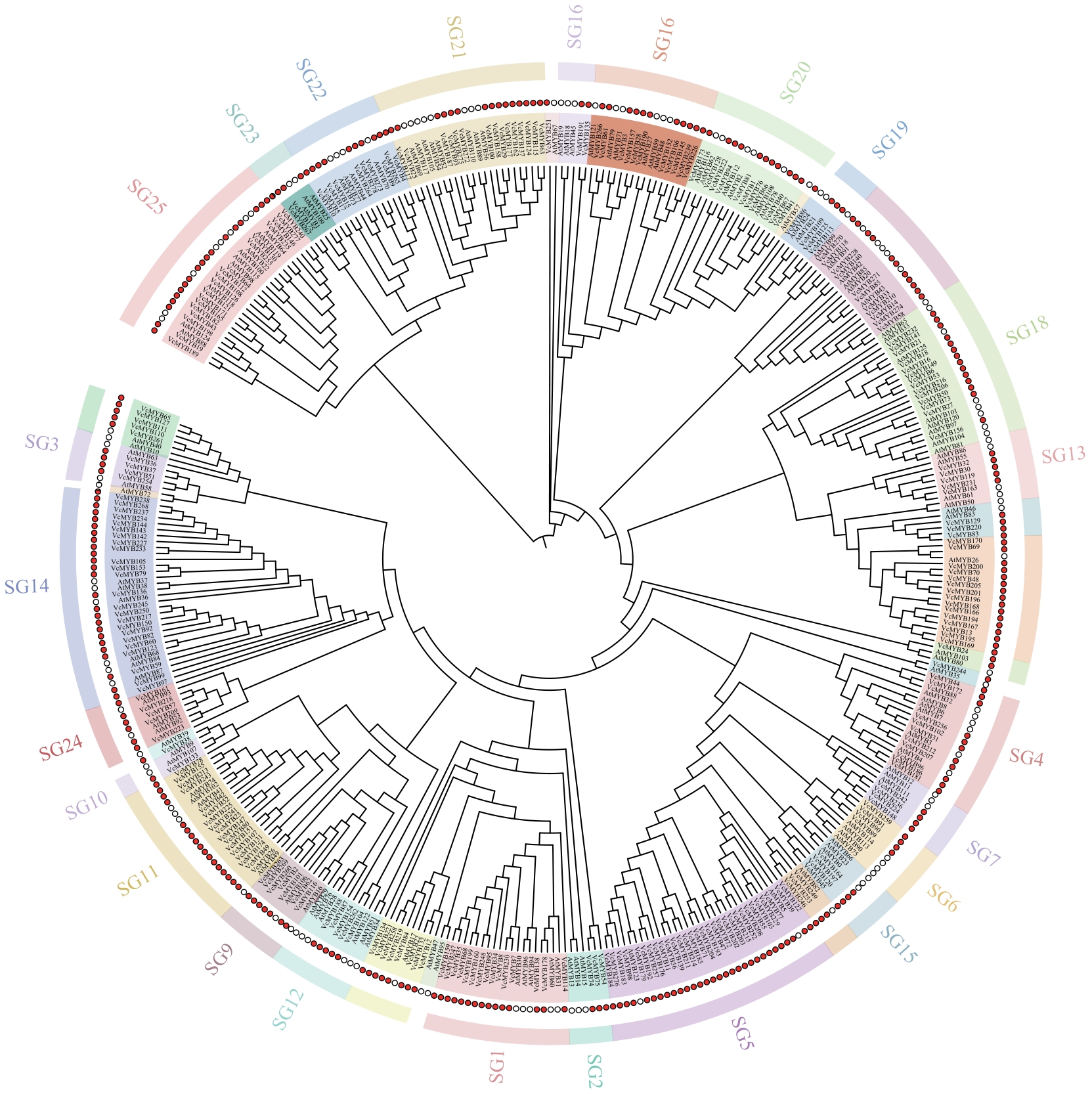
Fig. 1 Phylogenetic tree analysis of R2R3-MYB family in V. corymbosum and A. thalianaAt and Vc refer to genes from Arabidopsis thaliana and Vaccinium corymbosum, respectively; SG (Subgroup) refers to phylogenetically defined subgroups within the R2R3-MYB transcription factor family, the same below

Fig. 2 Analysis of gene structures, conserved motifs, and conserved domains in R2R3-MYB family members from blueberryA: Phylogenetic tree of blueberry R2R3-MYB family members; B: Gene structure of blueberry R2R3-MYB family members. C: Conserved motifs of blueberry R2R3-MYB family members. D: Conserved domains of blueberry R2R3-MYB family members. E: Sequence logo of conserved motifs in R2R3-MYB family members from blueberry

Fig. 5 Colinearity analysis of R2R3-MYB genes between blueberry and different speciesA: Colinearity analysis of R2R3-MYB genes between blueberry and different species (A. thaliana, Solanum lycopersicum, Vitis vinifera, Prunus persica, and Actinidia chinensis); B: The Venn diagram analyzed the same and different R2R3-MYB genes in blueberry and different species (A. thaliana, Solanum lycopersicum, Vitis vinifera, Prunus persica, and Actinidia chinensis)

Fig. 7 RT-qPCR detection of 14 R2R3-MYB family members in different tissues and different fruit development stages of blueberryA: Phenotypic maps of different tissues (roots, stems, leaves and flowers) and different fruit development stages (small green fruit, large green fruit, color-changing fruit and mature fruit), scale=0.25 cm. B: Relative expressions of 14 different R2R3-MYB genes in different tissues and different fruit development stages of blueberry. Data were analyzed by one-way ANOVA with Duncan's multiple range test, where different superscript letters denote significant differences (P<0.05)
| [1] | Plunkett BJ, Espley RV, Dare AP, et al. MYBA from blueberry (Vaccinium section Cyanococcus) is a subgroup 6 type R2R3MYB transcription factor that activates anthocyanin production [J]. Front Plant Sci, 2018, 9: 1300. |
| [2] | 李亚东, 裴嘉博, 陈丽, 等. 2020中国蓝莓产业年度报告 [J]. 吉林农业大学学报, 2021, 43(1): 1-8. |
| Li YD, Pei JB, Chen L, et al. China blueberry industry report 2020 [J]. J Jilin Agric Univ, 2021, 43(1): 1-8. | |
| [3] | 刘庆忠, 崔冬冬, 朱东姿. 世界及中国蓝莓产业现状 [J]. 落叶果树, 2024, 56(4): 1-7, 107. |
| Liu QZ, Cui DD, Zhu DZ. Current situation of blueberry industry in the world and China [J]. Deciduous Fruits, 2024, 56(4): 1-7, 107. | |
| [4] | Wen WW, Alseekh S, Fernie AR. Conservation and diversification of flavonoid metabolism in the plant Kingdom [J]. Curr Opin Plant Biol, 2020, 55: 100-108. |
| [5] | Jiang WB, Yin QG, Wu RR, et al. Role of a Chalcone isomerase-like protein in flavonoid biosynthesis in Arabidopsis thaliana [J]. J Exp Bot, 2015, 66(22): 7165-7179. |
| [6] | Miron A, Aprotosoaie AC, Trifan A, et al. Flavonoids as modulators of metabolic enzymes and drug transporters [J]. Ann N Y Acad Sci, 2017, 1398(1): 152-167. |
| [7] | Kuhn N, Guan L, Dai ZW, et al. Berry ripening: recently heard through the grapevine [J]. J Exp Bot, 2014, 65(16): 4543-4559. |
| [8] | Winkel-Shirley B. Biosynthesis of flavonoids and effects of stress [J]. Curr Opin Plant Biol, 2002, 5(3): 218-223. |
| [9] | Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana [J]. Curr Opin Plant Biol, 2001, 4(5): 447-456. |
| [10] | Kranz H, Scholz K, Weisshaar B. C-MYB oncogene-like genes encoding three MYB repeats occur in all major plant lineages [J]. Plant J, 2000, 21(2): 231-235. |
| [11] | Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis [J]. Trends Plant Sci, 2010, 15(10): 573-581. |
| [12] | Haga N, Kato K, Murase M, et al. R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana [J]. Development, 2007, 134(6): 1101-1110. |
| [13] | Zhu JTT, Choi RCY, Chu GKY, et al. Flavonoids possess neuroprotective effects on cultured pheochromocytoma PC12 cells: a comparison of different flavonoids in activating estrogenic effect and in preventing beta-amyloid-induced cell death [J]. J Agric Food Chem, 2007, 55(6): 2438-2445. |
| [14] | 冯盼盼, 陈鹏, 洪文杰, 等. 拟南芥MYB转录因子家族研究进展 [J]. 生命科学研究, 2016, 20(6): 555-560. |
| Feng PP, Chen P, Hong WJ, et al. Research progress of MYB transcription factor family in Arabidopsis thaliana [J]. Life Sci Res, 2016, 20(6): 555-560. | |
| [15] | Pandey A, Misra P, Choudhary D, et al. AtMYB12 expression in tomato leads to large scale differential modulation in transcriptome and flavonoid content in leaf and fruit tissues [J]. Sci Rep, 2015, 5: 12412. |
| [16] | Gonzalez A, Zhao MZ, Leavitt JM, et al. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings [J]. Plant J, 2008, 53(5): 814-827. |
| [17] | Wang XC, Wu J, Guan ML, et al. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis [J]. Plant J, 2020, 101(3): 637-652. |
| [18] | Baudry A, Heim MA, Dubreucq B, et al. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana [J]. Plant J, 2004, 39(3): 366-380. |
| [19] | An XH, Tian Y, Chen KQ, et al. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples [J]. Plant Cell Physiol, 2015, 56(4): 650-662. |
| [20] | Wang N, Xu HF, Jiang SH, et al. MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana) [J]. Plant J, 2017, 90(2): 276-292. |
| [21] | 李艳芳, 聂佩显, 张鹤华, 等. 蓝莓果实花青苷积累与内源激素含量动态变化 [J]. 北京林业大学学报, 2017, 39(2): 64-71. |
| Li YF, Nie PX, Zhang HH, et al. Dynamic changes of anthocyanin accumulation and endogenous hormone contents in blueberry [J]. J Beijing For Univ, 2017, 39(2): 64-71. | |
| [22] | Colle M, Leisner CP, Wai CM, et al. Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry [J]. GigaScience, 2019, 8(3): giz012. |
| [23] | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| [24] | Du H, Zhang L, Liu L, et al. Biochemical and molecular characterization of plant MYB transcription factor family [J]. Biochemistry, 2009, 74(1): 1-11. |
| [25] | Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family [J]. Plant Mol Biol, 1999, 41(5): 577-585. |
| [26] | Ruan MB, Guo X, Wang B, et al. Genome-wide characterization and expression analysis enables identification of abiotic stress-responsive MYB transcription factors in cassava (Manihot esculenta) [J]. J Exp Bot, 2017, 68(13): 3657-3672. |
| [27] | Matus JT, Aquea F, Arce-Johnson P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes [J]. BMC Plant Biol, 2008, 8: 83. |
| [28] | Zhao PP, Li Q, Li J, et al. Genome-wide identification and characterization of R2R3MYB family in Solanum lycopersicum [J]. Mol Genet Genomics, 2014, 289(6): 1183-1207. |
| [29] | Katiyar A, Smita S, Lenka SK, et al. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis [J]. BMC Genomics, 2012, 13: 544. |
| [30] | Wang HY, Zhai LL, Wang SW, et al. Identification of R2R3-MYB family in blueberry and its potential involvement of anthocyanin biosynthesis in fruits [J]. BMC Genomics, 2023, 24(1): 505. |
| [31] | Zapata L, Ding J, Willing EM, et al. Chromosome-level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms [J]. Proc Natl Acad Sci USA, 2016, 113(28): E4052-E4060. |
| [32] | Shi XY, Cao S, Wang X, et al. The complete reference genome for grapevine (Vitis viniferaL.) genetics and breeding [J]. Hortic Res, 2023, 10(5): uhad061. |
| [33] | Aoki K, Yano K, Suzuki A, et al. Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics [J]. BMC Genomics, 2010, 11: 210. |
| [34] | Wang WQ, Feng BX, Xiao JF, et al. Cassava genome from a wild ancestor to cultivated varieties [J]. Nat Commun, 2014, 5: 5110. |
| [35] | Yu J, Wang J, Lin W, et al. The genomes of Oryza sativa: a history of duplications [J]. PLoS Biol, 2005, 3(2): e38. |
| [36] | Jacquemin J, Ammiraju JSS, Haberer G, et al. Fifteen million years of evolution in the Oryza genus shows extensive gene family expansion [J]. Mol Plant, 2014, 7(4): 642-656. |
| [37] | Romani F, Reinheimer R, Florent SN, et al. Evolutionary history of homeodomain leucine zipper transcription factors during plant transition to land [J]. New Phytol, 2018, 219(1): 408-421. |
| [38] | Li YM, Kui LW, Liu Z, et al. Genome-wide analysis and expression profiles of the StR2R3-MYB transcription factor superfamily in potato (Solanum tuberosum L.) [J]. Int J Biol Macromol, 2020, 148: 817-832. |
| [39] | Du H, Feng BR, Yang SS, et al. The R2R3-MYB transcription factor gene family in maize [J]. PLoS One, 2012, 7(6): e37463. |
| [40] | Du H, Yang SS, Liang Z, et al. Genome-wide analysis of the MYB transcription factor superfamily in soybean [J]. BMC Plant Biol, 2012, 12: 106. |
| [41] | Wang YS, Nie F, Shahid MQ, et al. Molecular footprints of selection effects and whole genome duplication (WGD) events in three blueberry species: detected by transcriptome dataset [J]. BMC Plant Biol, 2020, 20(1): 250. |
| [42] | Li WB, Ding ZH, Ruan MB, et al. Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthesis [J]. Sci Rep, 2017, 7(1): 16861. |
| [43] | Hou HL, Zhang CW, Hou XL. Cloning and functional analysis of BcMYB101 gene involved in leaf development in pak choi (Brassica rapa ssp. Chinensis) [J]. Int J Mol Sci, 2020, 21(8): 2750. |
| [44] | Stracke R, Ishihara H, Huep G, et al. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling [J]. Plant J, 2007, 50(4): 660-677. |
| [45] | Czemmel S, Stracke R, Weisshaar B, et al. The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries [J]. Plant Physiol, 2009, 151(3): 1513-1530. |
| [46] | Yao PF, Huang YJ, Dong QX, et al. FtMYB6, a light-induced SG7 R2R3-MYB transcription factor, promotes flavonol biosynthesis in Tartary buckwheat (Fagopyrum tataricum) [J]. J Agric Food Chem, 2020, 68(47): 13685-13696. |
| [47] | Chen Q, Liu XJ, Hu YY, et al. Transcriptomic profiling of fruit development in black raspberry Rubus coreanus [J]. Int J Genomics, 2018, 2018: 8084032. |
| [48] | Zhang L, Duan ZD, Ma S, et al. SlMYB7, an AtMYB4-like R2R3-MYB transcription factor, inhibits anthocyanin accumulation in Solanum lycopersicum fruits [J]. J Agric Food Chem, 2023, 71(48): 18758-18768. |
| [49] | Jiang LY, Yue ML, Liu YQ, et al. A novel R2R3-MYB transcription factor FaMYB5 positively regulates anthocyanin and proanthocyanidin biosynthesis in cultivated strawberries (Fragaria × Ananassa) [J]. Plant Biotechnol J, 2023, 21(6): 1140-1158. |
| [50] | Lai B, Du LN, Hu B, et al. Characterization of a novel Litchi R2R3-MYB transcription factor that involves in anthocyanin biosynthesis and tissue acidification [J]. BMC Plant Biol, 2019, 19(1): 62. |
| [51] | Qi XL, Liu CL, Song LL, et al. A sweet cherry glutathione S-transferase gene, PavGST1, plays a central role in fruit skin coloration [J]. Cells, 2022, 11(7): 1170. |
| [52] | Zhou H, Kui LW, Wang FR, et al. Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation [J]. New Phytol, 2019, 221(4): 1919-1934. |
| [1] | DONG Xiang-xiang, MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song. Bioinformatics Analysis and Flowering Regulation Function of FveBBX20 Gene in Woodland Strawberry [J]. Biotechnology Bulletin, 2025, 41(9): 115-123. |
| [2] | LA Gui-xiao, ZHAO Yu-long, DAI Dan-dan, YU Yong-liang, GUO Hong-xia, SHI Gui-xia, JIA Hui, YANG Tie-gang. Identification of Plasma Membrane H+-ATPase Gene Family in Safflower and Expression Analysis in Response to Low Nitrogen and Low Phosphorus Stress [J]. Biotechnology Bulletin, 2025, 41(8): 220-233. |
| [3] | LAI Shi-yu, LIANG Qiao-lan, WEI Lie-xin, NIU Er-bo, CHEN Ying-e, ZHOU Xin, YANG Si-zheng, WANG Bo. The Role of NbJAZ3 in the Infection of Nicotiana benthamiana by Alfalfa Mosaic Virus [J]. Biotechnology Bulletin, 2025, 41(8): 186-196. |
| [4] | LI Kai-yue, DENG Xiao-xia, YIN Yuan, DU Ya-tong, XU Yuan-jing, WANG Jing-hong, YU Song, LIN Ji-xiang. Identification of LEA Gene Family and Analysis on Its Response to Aluminum Stress in Ricinus communis L. [J]. Biotechnology Bulletin, 2025, 41(7): 128-138. |
| [5] | GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant [J]. Biotechnology Bulletin, 2025, 41(7): 214-225. |
| [6] | QU Mei-ling, ZHOU Si-min, ZHANG Jing-yu, HE Jia-wei, ZHU Jia-yuan, LIU Xiao-rong, TONG Qiao-zhen, ZHOU Ri-bao, LIU Xiang-dan. Identification and Expression Analysis of bHLH Transcription Gene Family in Lonicera macranthoides [J]. Biotechnology Bulletin, 2025, 41(6): 256-268. |
| [7] | ZHAO Jing, GUO Qian, LI Rui-qi, LEI Ying-yang, YUE Ai-qin, ZHAO Jin-zhong, YIN Cong-cong, DU Wei-jun, NIU Jing-ping. Sequence Analysis and Induced Expression Analysis of GmGST Gene Cluster Genes in Soybean [J]. Biotechnology Bulletin, 2025, 41(5): 129-140. |
| [8] | LI Zhi-qiang, LUO Zheng-qian, XU Lin-li, ZHOU Guo-hui, QU Si-yu, LIU En-liang, XU Dong-ting. Identification of the Soybean R2R3-MYB Gene Family Based on the T2T Genome and Their Expression Analysis under Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(5): 141-152. |
| [9] | YANG Chun, WANG Xiao-qian, WANG Hong-jun, CHAO Yue-hui. Cloning, Subcellular Localization and Expression Analysis of MtZHD4 Gene from Medicago truncatula [J]. Biotechnology Bulletin, 2025, 41(5): 244-254. |
| [10] | TIAN Qin, LIU Kui, WU Xiang-wei, JI Yuan-yuan, CAO Yi-bo, ZHANG Ling-yun. Functional Study of Transcription Factor VcMYB17 in Regulating Drought Tolerance in Blueberry [J]. Biotechnology Bulletin, 2025, 41(4): 198-210. |
| [11] | LIU Tao, WANG Zhi-qi, WU Wen-bo, SHI Wen-ting, WANG Chao-nan, DU Chong, YANG Zhong-min. Identification and Expression Analysis of the GRAM Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(4): 145-155. |
| [12] | WANG Tian-xi, YANG Bing-song, PAN Rong-jun, GAI Wen-xian, LIANG Mei-xia. Identification of the Apple PLATZ Gene Family and Functional Study of the MdPLATZ9 Gene [J]. Biotechnology Bulletin, 2025, 41(4): 176-187. |
| [13] | BAN Qiu-yan, ZHAO Xin-yue, CHI Wen-jing, LI Jun-sheng, WANG Qiong, XIA Yao, LIANG Li-yun, HE Wei, LI Ye-yun, ZHAO Guang-shan. Cloning of Phytochrome Interaction Factor CsPIF3a and Its Response to Light and Temperature Stress in Camellia sinensis [J]. Biotechnology Bulletin, 2025, 41(4): 256-265. |
| [14] | LU Yong-jie, XIA Hai-qian, LI Yong-ling, ZHANG Wen-jian, YU Jing, ZHAO Hui-na, WANG Bing, XU Ben-bo, LEI Bo. Cloning and Expression Analysis of AP2/ERF Transcription Factor NtESR2 in Nicotiana tabacum [J]. Biotechnology Bulletin, 2025, 41(4): 266-277. |
| [15] | ZHANG Yi-xuan, MA Yu, WANG Tong-tong, SHENG Su-ao, SONG Jia-feng, LYU Zhao-yan, ZHU Xiao-biao, HOU Hua-lan. Genome-wide Identification and Expression Profiles of DIR Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(3): 123-136. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||