








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (9): 207-218.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0218
WANG Bin1,2,3( ), LIN Chong2, YUAN Xiao2,4, JIANG Yuan-yuan1,2,3, WANG Yu-kun1,2,3, XIAO Yan-hui1,2,3(
), LIN Chong2, YUAN Xiao2,4, JIANG Yuan-yuan1,2,3, WANG Yu-kun1,2,3, XIAO Yan-hui1,2,3( )
)
Received:2025-03-02
Online:2025-09-26
Published:2025-09-24
Contact:
XIAO Yan-hui
E-mail:b_wang@sgu.edu.cn;yhxiao@sgu.edu.cn
WANG Bin, LIN Chong, YUAN Xiao, JIANG Yuan-yuan, WANG Yu-kun, XIAO Yan-hui. Cloning of bHLH Transcription Factor UNE10 and Its Regulatory Roles in the Biosynthesis of Volatile Compounds in Clove Basil[J]. Biotechnology Bulletin, 2025, 41(9): 207-218.
基因名称 Gene name | 正向序列 Forward sequence(5′-3′) | 反向序列 Reverse sequence(5′-3′) | 实验用途 Experimental purpose |
|---|---|---|---|
| OgUNE10 | ggactctagaggatccccgggATGAAGACATTGCAGAAGTTGGTC | ataagggactgaccacccgggGTTCTTG GAGGCAGAAGCCG | TA克隆、亚细胞定位和过表达 TA cloning, subcellular localization, and overexpression |
| OgACTIN7 | GGAGCTCGTCTTTGCTGTCT | GAGCGGGAAATTGTGAGGGA | RT-qPCR |
| OgUNE10 | TGTCTATGATGGGCATGGGC | GCCTGACAAGCAAAGAAGGC | |
| OgPAL1 | TTTTTCGCCCGTCAAGAAGC | CTGGAGAGTGGGAATCCAGC | |
| OgPAL2 | GGATCCCCTGCAGAAACCAA | ATCAATCAGCGGGTTGTCGT | |
| OgPAL4 | ATGCCGGAGAAACCTTGGAG | ATTCTCCCTCGCTCAGCAAC | |
| OgMES1 | GATAGGCCCTCGTGGAGTTG | TAACTGCATCAACGCCCAGT | |
| OgMES2 | CTCCATGGTCCGGGCTATTC | TGTCCCCTCAAATCCAAGGC | |
| OgMAT1 | TCTACACACGCACACTCGAC | GTACCAGCACCATGCTCCAT | |
| OgEFS1 | CCACACTTGTTGCTCCTTGC | GCTGGTAGTTTCCAGAGCGT | |
| OgMD1 | GGTCTTCAACAGCCCGATCA | ACGTCGGAACTTTTGACGGA | |
| OgCyt P450 | AATCCGATAATGGGTCCGCC | TTGCTGCAGGAGAAGTAGCC | |
| OgG8H1 | TGGGTCCGTCAAATCCTTGG | ACTGCCGGAAAAGTAGCCTC | |
| OgiPR1 | GCAAAGTGAAGAGGCCAACG | TTCTGCCGAATGAATGGGCT | |
| OgGD2 | ATCTTGGCGTGGACGAGTTT | TCAGCAACGGAACAATGGGA |
Table 1 Primer sequence
基因名称 Gene name | 正向序列 Forward sequence(5′-3′) | 反向序列 Reverse sequence(5′-3′) | 实验用途 Experimental purpose |
|---|---|---|---|
| OgUNE10 | ggactctagaggatccccgggATGAAGACATTGCAGAAGTTGGTC | ataagggactgaccacccgggGTTCTTG GAGGCAGAAGCCG | TA克隆、亚细胞定位和过表达 TA cloning, subcellular localization, and overexpression |
| OgACTIN7 | GGAGCTCGTCTTTGCTGTCT | GAGCGGGAAATTGTGAGGGA | RT-qPCR |
| OgUNE10 | TGTCTATGATGGGCATGGGC | GCCTGACAAGCAAAGAAGGC | |
| OgPAL1 | TTTTTCGCCCGTCAAGAAGC | CTGGAGAGTGGGAATCCAGC | |
| OgPAL2 | GGATCCCCTGCAGAAACCAA | ATCAATCAGCGGGTTGTCGT | |
| OgPAL4 | ATGCCGGAGAAACCTTGGAG | ATTCTCCCTCGCTCAGCAAC | |
| OgMES1 | GATAGGCCCTCGTGGAGTTG | TAACTGCATCAACGCCCAGT | |
| OgMES2 | CTCCATGGTCCGGGCTATTC | TGTCCCCTCAAATCCAAGGC | |
| OgMAT1 | TCTACACACGCACACTCGAC | GTACCAGCACCATGCTCCAT | |
| OgEFS1 | CCACACTTGTTGCTCCTTGC | GCTGGTAGTTTCCAGAGCGT | |
| OgMD1 | GGTCTTCAACAGCCCGATCA | ACGTCGGAACTTTTGACGGA | |
| OgCyt P450 | AATCCGATAATGGGTCCGCC | TTGCTGCAGGAGAAGTAGCC | |
| OgG8H1 | TGGGTCCGTCAAATCCTTGG | ACTGCCGGAAAAGTAGCCTC | |
| OgiPR1 | GCAAAGTGAAGAGGCCAACG | TTCTGCCGAATGAATGGGCT | |
| OgGD2 | ATCTTGGCGTGGACGAGTTT | TCAGCAACGGAACAATGGGA |
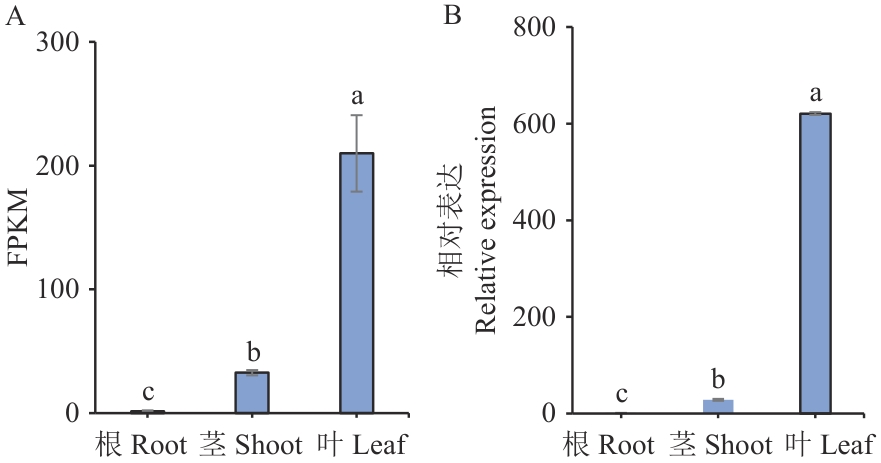
Fig. 1 Expressions of OgUNE10 in different tissues of clove basil (Ocimum gratissimum)FPKM refers to the number of reads per kilobase length from alignment to a gene per million reads, error bars indicate standard errors. The same below
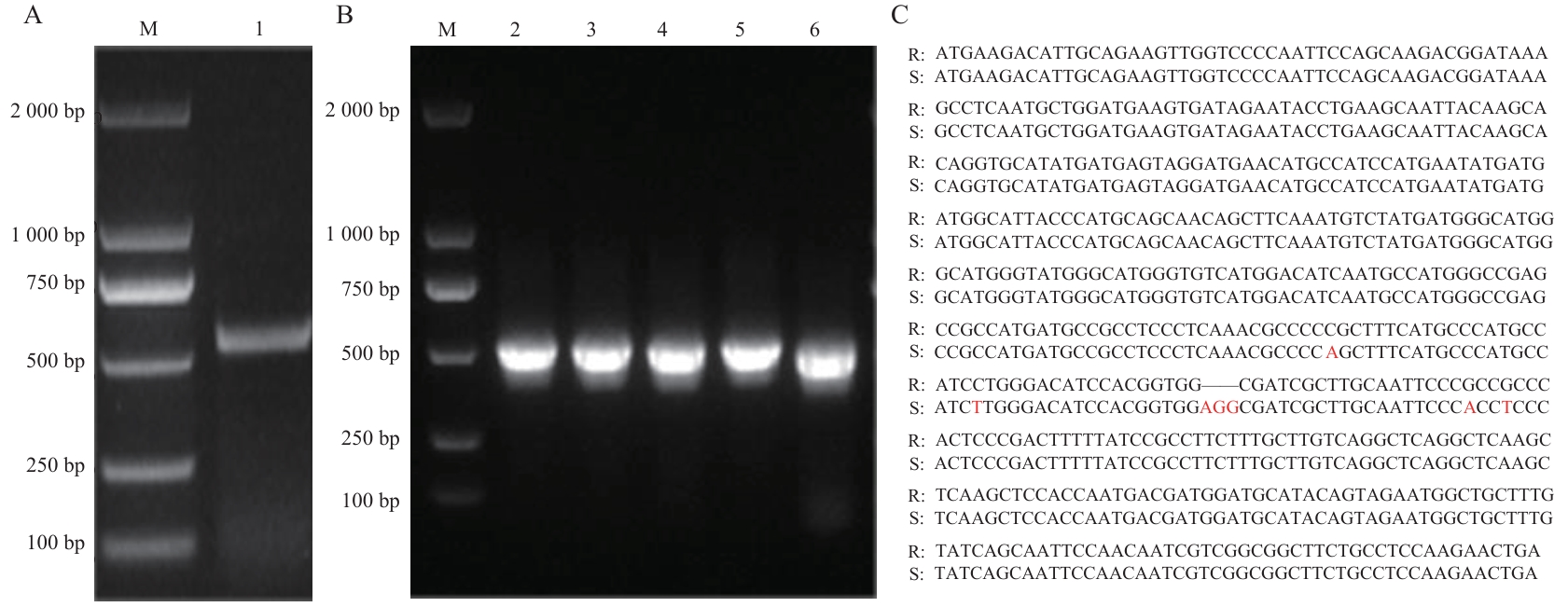
Fig. 2 Electrophoretic image of OgUNE10 PCR products and sequencesA: Electrophoretic image of PCR product amplified using cDNA as template. B: Electrophoretic image of PCR products amplified using the recombinant plasmid linking the full length of OgUNE10 as templates. C: Full length of OgUNE10 assembled by RNA-Seq and obtained from cloning sequence. Lane M: DNA marker. Lanes 1-6: Full length OgUNE10 PCR amplified products. R: RNA-Seq assembled sequence. S: Sanger sequencing sequence; red letters are different bases
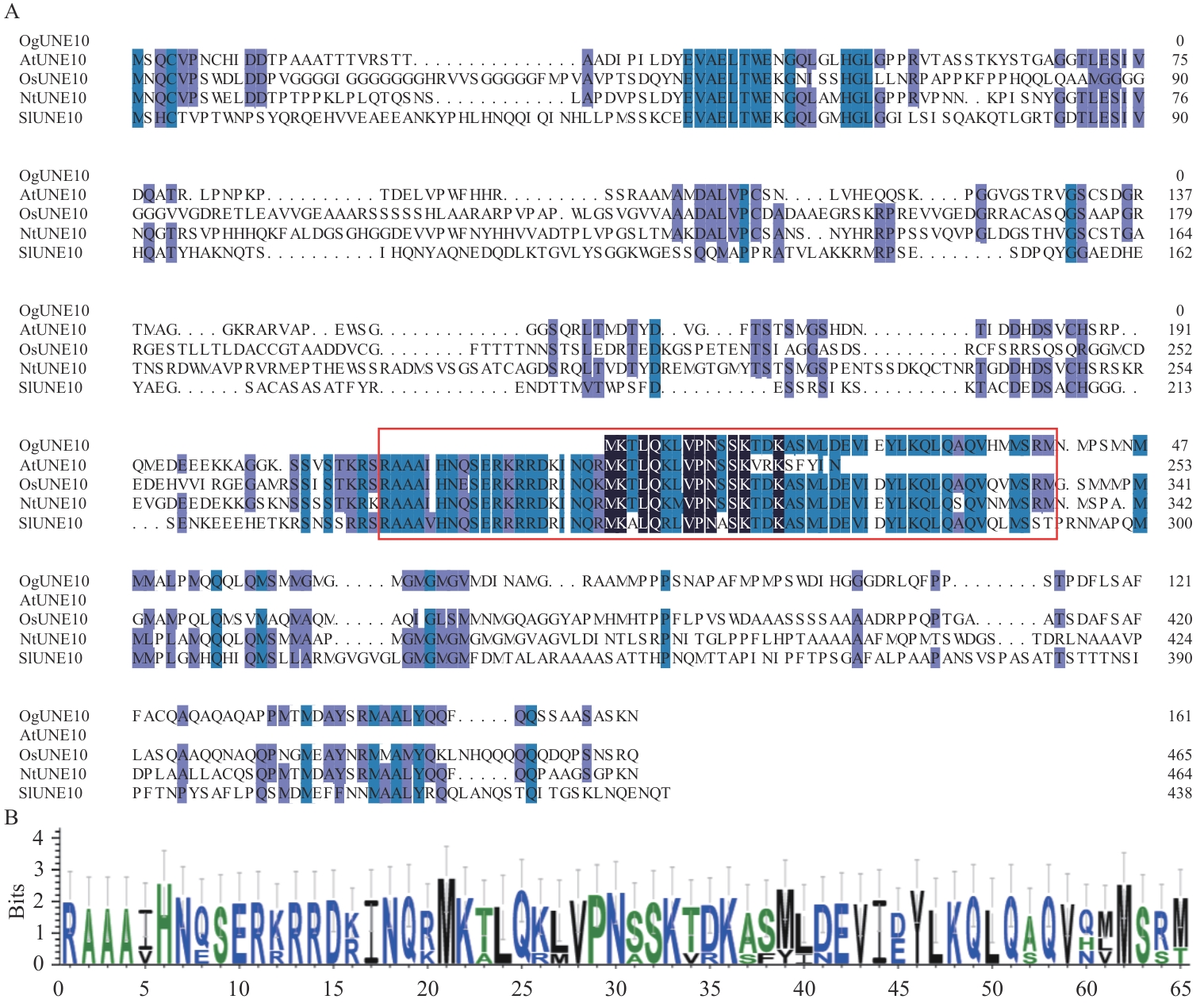
Fig. 3 Multiple sequence alignment (A) and conservation analysis (B) of OgUNE10Sequences in red box is the bHLH conserved domain, protein accession No. are indicated in Fig. 4-B below
| 植物类型Plant species | 丁香罗勒Ocimum gratissimum | 水稻Oryza sativa | 拟南芥Arabidopsis thaliana | 烟草Nicotiana tabacum | 番茄Solanum lycopersicum |
|---|---|---|---|---|---|
| 丁香罗勒 Ocimum gratissimum | 100 | ||||
| 水稻 Oryza sativa | 23.55 | 100 | |||
| 拟南芥 Arabidopsis thaliana | 29.51 | 52.57 | 100 | ||
| 烟草 Nicotiana tabacum | 24.31 | 47.90 | 52.89 | 100 | |
| 番茄 Solanum lycopersicum | 22.87 | 47.98 | 53.05 | 79.45 | 100 |
Table 2 Similarity comparison of the sequences between OgUNE10 of clove basil and UNE10 proteins from four model plants/%
| 植物类型Plant species | 丁香罗勒Ocimum gratissimum | 水稻Oryza sativa | 拟南芥Arabidopsis thaliana | 烟草Nicotiana tabacum | 番茄Solanum lycopersicum |
|---|---|---|---|---|---|
| 丁香罗勒 Ocimum gratissimum | 100 | ||||
| 水稻 Oryza sativa | 23.55 | 100 | |||
| 拟南芥 Arabidopsis thaliana | 29.51 | 52.57 | 100 | ||
| 烟草 Nicotiana tabacum | 24.31 | 47.90 | 52.89 | 100 | |
| 番茄 Solanum lycopersicum | 22.87 | 47.98 | 53.05 | 79.45 | 100 |
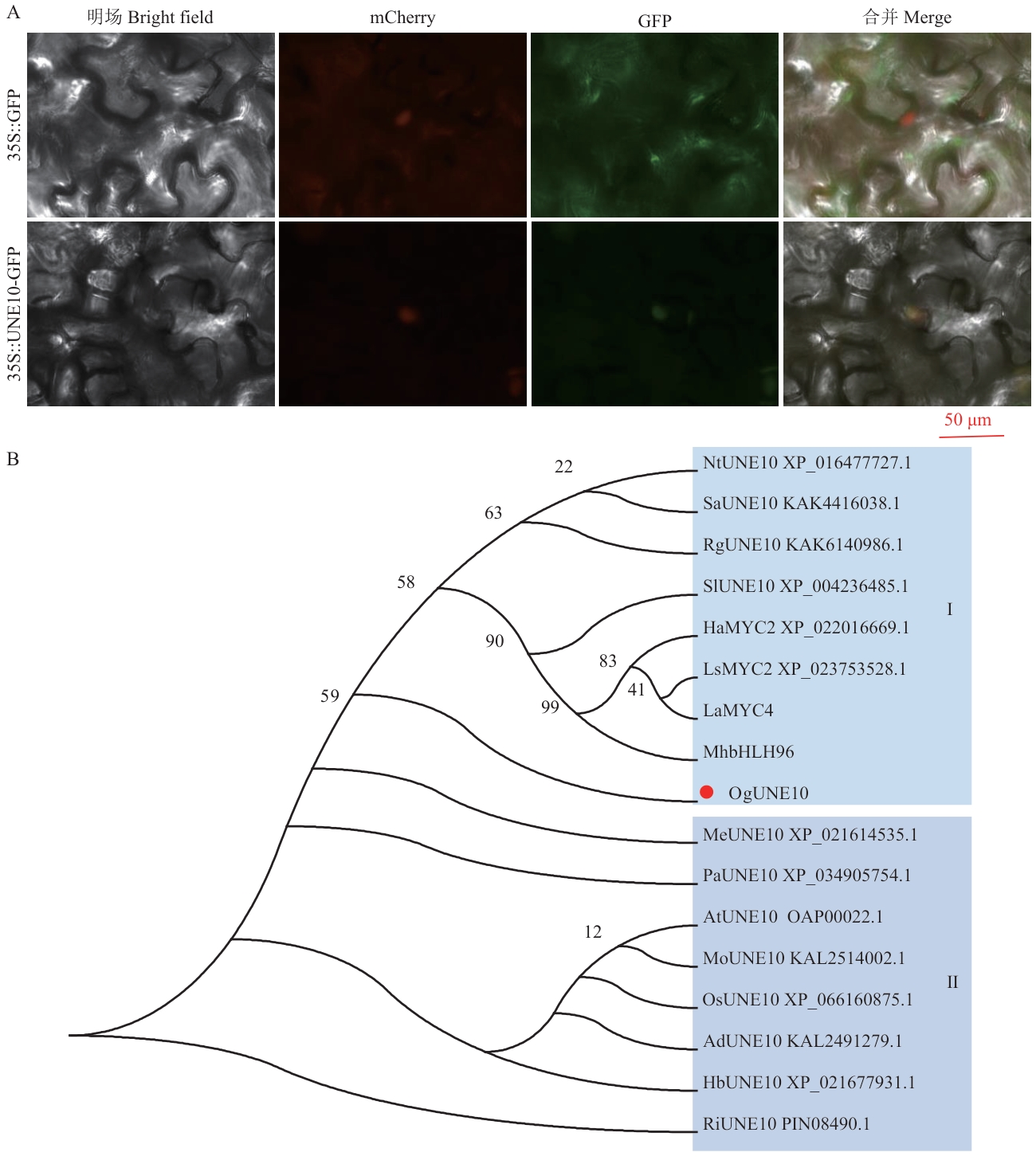
Fig. 4 Subcellular localization (A) and phylogenetic analysis (B) of OgUNE10 proteinThe amino acid sequences of LaMYC4 and MhbHLH96 can be found in the references [13, 26], respectively
序号 No. | 化合物 Compound | 保留时间 Retention time (min) | 相对含量 Relative content (%) | |
|---|---|---|---|---|
| 35S::GFP | 35S::OgUNE10-GFP | |||
| 1 | 3-己烯醛 3-Hexenal | 4.05 | 0.27±0.01 | 0.38±0.08* |
| 2 | α-蒎烯 α-Pinene | 6.19 | 0.23±0.04 | 0.25±0.04 |
| 3 | 莰烯 Camphene | 6.45 | - | 0.10±0.01 |
| 4 | 香桧烯 Sabinene | 6.84 | 0.13±0.03 | - |
| 5 | β-蒎烯 β-Pinene | 6.90 | 0.24±0.04 | 0.27±0.02 |
| 6 | β-月桂烯 β-Myrcene | 7.08 | 0.14±0.01 | 0.19±0.03 |
| 7 | D-枞油烯 D-sylvestrene | 7.73 | 0.10±0.01 | 0.15±0.01* |
| 8 | 桉叶油醇 Eucalyptol | 7.78 | 0.64±0.08 | 0.71±0.06 |
| 9 | 反式-β-罗勒烯 Trans-β-Ocimene | 8.00 | 0.77±0.03 | 0.74±0.03 |
| 10 | 茴香酮 Fenchone | 8.71 | 0.23±0.04 | 1.02±0.05* |
| 11 | 芳樟醇 Linalool | 8.81 | 1.53±0.05 | 4.65±0.19* |
| 12 | (+)-2-莰酮 (+)-2-Bornanone | 9.60 | 0.28±0.03 | 0.71±0.12* |
| 13 | L-4-松油醇 L-terpinen-4-ol | 10.06 | 0.21±0.03* | 0.14±0.03 |
| 14 | L-α-松油醇 L-α-Terpineol | 10.25 | 0.09±0.01 | 0.10±0.02 |
| 15 | 草蒿脑 Estragole | 10.35 | 17.43±1.33* | 1.08±0.03 |
| 16 | 顺式肉桂酸甲酯 Methyl cis-cinnamate | 11.87 | 0.17±0.05 | - |
| 17 | 丁香酚 Eugenol | 12.59 | 20.10±5.75 | 46.58±1.11* |
| 18 | 肉桂酸甲酯 Methyl cinnamate | 12.94 | 51.39±5.28* | 34.43±0.28 |
| 19 | β-榄香烯 β-Elemene | 13.09 | 0.13±0.01 | 0.28±0.01* |
| 20 | 甲基丁香酚 Methyl eugenol | 13.16 | 1.48±0.23 | 3.73±0.04* |
| 21 | 反式-α-佛手柑油烯 Trans-α-Bergamotene | 13.64 | 0.84±0.09 | 3.73±0.04* |
| 22 | α-愈创木烯 α-Guaiene | 13.71 | - | 0.09±0.01 |
| 23 | 蛇麻烯 Humulene | 13.95 | 0.21±0.05 | 0.35±0.03* |
| 24 | 顺式-4(15),5-衣兰油二烯 Cis-Muurola-4(15),5-diene | 14.06 | 0.19±0.02 | 0.19±0.04 |
| 25 | 大根香叶烯D Germacrene D | 14.29 | 0.95±0.11 | 0.97±0.06 |
| 26 | β-环大根香叶烷 β-Cyclogermacrane | 14.48 | 0.22±0.02 | 0.33±0.02* |
| 27 | δ-愈创木烯 δ-Guaiene | 14.58 | - | 0.14±0.02 |
| 28 | γ-愈创木烯 γ-Cadinene | 14.68 | 0.62±0.03 | 0.83±0.04* |
| 29 | τ-杜松醇 τ-Cadinol | 16.18 | 0.83±0.08 | 0.77±0.06 |
| 总鉴定率 Total identification rate (%) | 99.42 | 99.49 | ||
Table 3 Effects of OgUNE10 overexpressionon the types and relative content of volatile compounds in clove basil leaves
序号 No. | 化合物 Compound | 保留时间 Retention time (min) | 相对含量 Relative content (%) | |
|---|---|---|---|---|
| 35S::GFP | 35S::OgUNE10-GFP | |||
| 1 | 3-己烯醛 3-Hexenal | 4.05 | 0.27±0.01 | 0.38±0.08* |
| 2 | α-蒎烯 α-Pinene | 6.19 | 0.23±0.04 | 0.25±0.04 |
| 3 | 莰烯 Camphene | 6.45 | - | 0.10±0.01 |
| 4 | 香桧烯 Sabinene | 6.84 | 0.13±0.03 | - |
| 5 | β-蒎烯 β-Pinene | 6.90 | 0.24±0.04 | 0.27±0.02 |
| 6 | β-月桂烯 β-Myrcene | 7.08 | 0.14±0.01 | 0.19±0.03 |
| 7 | D-枞油烯 D-sylvestrene | 7.73 | 0.10±0.01 | 0.15±0.01* |
| 8 | 桉叶油醇 Eucalyptol | 7.78 | 0.64±0.08 | 0.71±0.06 |
| 9 | 反式-β-罗勒烯 Trans-β-Ocimene | 8.00 | 0.77±0.03 | 0.74±0.03 |
| 10 | 茴香酮 Fenchone | 8.71 | 0.23±0.04 | 1.02±0.05* |
| 11 | 芳樟醇 Linalool | 8.81 | 1.53±0.05 | 4.65±0.19* |
| 12 | (+)-2-莰酮 (+)-2-Bornanone | 9.60 | 0.28±0.03 | 0.71±0.12* |
| 13 | L-4-松油醇 L-terpinen-4-ol | 10.06 | 0.21±0.03* | 0.14±0.03 |
| 14 | L-α-松油醇 L-α-Terpineol | 10.25 | 0.09±0.01 | 0.10±0.02 |
| 15 | 草蒿脑 Estragole | 10.35 | 17.43±1.33* | 1.08±0.03 |
| 16 | 顺式肉桂酸甲酯 Methyl cis-cinnamate | 11.87 | 0.17±0.05 | - |
| 17 | 丁香酚 Eugenol | 12.59 | 20.10±5.75 | 46.58±1.11* |
| 18 | 肉桂酸甲酯 Methyl cinnamate | 12.94 | 51.39±5.28* | 34.43±0.28 |
| 19 | β-榄香烯 β-Elemene | 13.09 | 0.13±0.01 | 0.28±0.01* |
| 20 | 甲基丁香酚 Methyl eugenol | 13.16 | 1.48±0.23 | 3.73±0.04* |
| 21 | 反式-α-佛手柑油烯 Trans-α-Bergamotene | 13.64 | 0.84±0.09 | 3.73±0.04* |
| 22 | α-愈创木烯 α-Guaiene | 13.71 | - | 0.09±0.01 |
| 23 | 蛇麻烯 Humulene | 13.95 | 0.21±0.05 | 0.35±0.03* |
| 24 | 顺式-4(15),5-衣兰油二烯 Cis-Muurola-4(15),5-diene | 14.06 | 0.19±0.02 | 0.19±0.04 |
| 25 | 大根香叶烯D Germacrene D | 14.29 | 0.95±0.11 | 0.97±0.06 |
| 26 | β-环大根香叶烷 β-Cyclogermacrane | 14.48 | 0.22±0.02 | 0.33±0.02* |
| 27 | δ-愈创木烯 δ-Guaiene | 14.58 | - | 0.14±0.02 |
| 28 | γ-愈创木烯 γ-Cadinene | 14.68 | 0.62±0.03 | 0.83±0.04* |
| 29 | τ-杜松醇 τ-Cadinol | 16.18 | 0.83±0.08 | 0.77±0.06 |
| 总鉴定率 Total identification rate (%) | 99.42 | 99.49 | ||
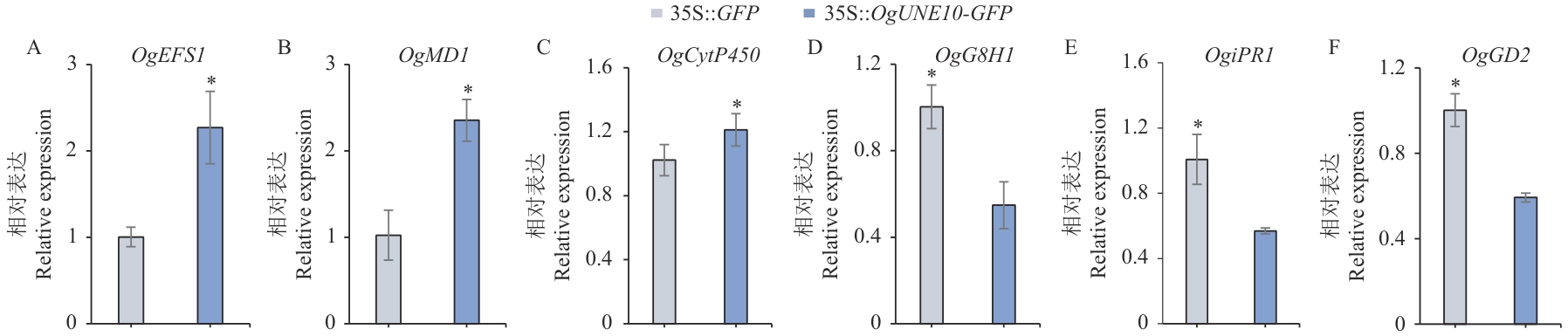
Fig. 8 Expression analysis of genes related to terpenoids biosynthesisOgEFS1: (-)-Endo-fenchol synthase 1. OgMD1: Mannitol dehydrogenase 1. OgCyt P450: Cytochrome P450. OgG8H1: Geraniol 8-hydroxylase 1. OgiPR1: (-)-isopiperitenone reductase 1. OgGD2: Geraniol dehydrogenase 2
| [1] | de Sousa DP, Damasceno ROS, Amorati R, et al. Essential oils: chemistry and pharmacological activities [J]. Biomolecules, 2023, 13(7): 1144. |
| [2] | Tian F, Woo SY, Lee SY, et al. Antifungal activity of essential oil and plant-derived natural compounds against Aspergillus flavus [J]. Antibiotics, 2022, 11(12): 1727. |
| [3] | Hudz N, Kobylinska L, Pokajewicz K, et al. Mentha piperita: essential oil and extracts, their biological activities, and perspectives on the development of new medicinal and cosmetic products [J]. Molecules, 2023, 28(21): 7444. |
| [4] | He JM, Hadidi M, Yang SY, et al. Natural food preservation with ginger essential oil: Biological properties and delivery systems [J]. Food Res Int, 2023, 173(Pt 1): 113221. |
| [5] | Tian YQ, Zhou L, Liu JP, et al. Metal-organic frameworks-based moisture responsive essential oil hydrogel beads for fresh-cut pineapple preservation [J]. Food Chem, 2024, 451: 139440. |
| [6] | Zhang WL, Pan YG, Jiang YM, et al. Advances in gas fumigation technologies for postharvest fruit preservation [J]. Crit Rev Food Sci Nutr, 2024, 64(24): 8689-8708. |
| [7] | Dinday S, Ghosh S. Recent advances in triterpenoid pathway elucidation and engineering [J]. Biotechnol Adv, 2023, 68: 108214. |
| [8] | Rinaldi MA, Ferraz CA, Scrutton NS. Alternative metabolic pathways and strategies to high-titre terpenoid production in Escherichia coli [J]. Nat Prod Rep, 2022, 39(1): 90-118. |
| [9] | Song L, Li WT, Chen XW. Transcription factor is not just a transcription factor [J]. Trends Plant Sci, 2022, 27(11): 1087-1089. |
| [10] | Gao F, Dubos C. The Arabidopsis bHLH transcription factor family [J]. Trends Plant Sci, 2024, 29(6): 668-680. |
| [11] | 李宇, 李素贞, 陈茹梅, 等. 植物bHLH转录因子调控铁稳态的研究进展 [J]. 生物技术通报, 2023, 39(7): 26-36. |
| Li Y, Li SZ, Chen RM, et al. Advances in the regulation of iron homeostasis by bHLH transcription factors in plant [J]. Biotechnol Bull, 2023, 39(7): 26-36. | |
| [12] | 安昌, 陆琳, 沈梦千, 等. 植物bHLH基因家族研究进展及在药用植物中的应用前景 [J]. 生物技术通报, 2023, 39(10): 1-16. |
| An C, Lu L, Shen MQ, et al. Research progress of bHLH gene family in plants and its application prospects in medical plants [J]. Biotechnol Bull, 2023, 39(10): 1-16. | |
| [13] | 王斌, 袁晓, 蒋园园, 等. bHLH96的克隆及其在薄荷萜烯生物合成调控中的功能 [J]. 生物技术通报, 2024, 40(1): 281-293. |
| Wang B, Yuan X, Jiang YY, et al. Cloning of bHLH96 gene and its roles in regulating the biosynthesis of peppermint terpenes [J]. Biotechnol Bull, 2024, 40(1): 281-293. | |
| [14] | He ZH, Wang ZB, Nie XG, et al. UNFERTILIZED EMBRYO SAC 12 phosphorylation plays a crucial role in conferring salt tolerance [J]. Plant Physiol, 2022, 188(2): 1385-1401. |
| [15] | Lu SF, Li QZ, Wei HR, et al. Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa [J]. Proc Natl Acad Sci USA, 2013, 110(26): 10848-10853. |
| [16] | Song CW, Guo YY, Shen WW, et al. PagUNE12 encodes a basic helix-loop-helix transcription factor that regulates the development of secondary vascular tissue in poplar [J]. Plant Physiol, 2023, 192(2): 1046-1062. |
| [17] | Vieira RF, Grayer RJ, Paton A, et al. Genetic diversity of Ocimum gratissimum L. based on volatile oil constituents, flavonoids and RAPD markers [J]. Biochem Syst Ecol, 2001, 29(3): 287-304. |
| [18] | 刘腾浩, 姜子涛, 李荣. 天然调味香料丁香罗勒精油的研究进展 [J]. 中国调味品, 2013, 38(7): 113-116. |
| Liu TH, Jiang ZT, Li R. Research progress on essential oil of Ocimum gratissimum L [J]. China Condiment, 2013, 38(7): 113-116. | |
| [19] | Ugbogu OC, Emmanuel O, Agi GO, et al. A review on the traditional uses, phytochemistry, and pharmacological activities of clove basil (Ocimum gratissimum L.) [J]. Heliyon, 2021, 7(11): e08404. |
| [20] | 张瑜, 何聪芬. 不同品种罗勒精油的成分及其功效概况 [J]. 香料香精化妆品, 2021(6): 81-88. |
| Zhang Y, He CF. Overview of composition and efficacy of different basil essential oils [J]. Flavour Fragr Cosmet, 2021(6): 81-88. | |
| [21] | 杜丽霞, 占豪, 姜子涛. 调味香料丁香罗勒精油成分、抗氧化及抑菌活性 [J]. 中国调味品, 2021, 46(4): 174-178, 182. |
| Du LX, Zhan H, Jiang ZT. The components, antioxidant and antibacterial activities of essential oil of flavoring spice Ocimum gratissimum L. [J]. China Cond, 2021, 46(4): 174-178, 182. | |
| [22] | 王斌, 王玉昆, 肖艳辉. 丁香罗勒(Ocimum gratissimum)叶片响应镉胁迫的比较转录组学分析 [J]. 生物技术通报, 2025, 41(3): 255-270. |
| Wang B, Wang YK, Xiao YH. Comparative transcriptomic analysis of clove basil (Ocimum gratissimum) leaves in response to cadmium stress [J]. Biotechnol Bull, 2025, 41(3): 255-270. | |
| [23] | Wang HX, Ren J, Zhou SY, et al. Molecular regulation of oil gland development and biosynthesis of essential oils in Citrus spp [J]. Science, 2024, 383(6683): 659-666. |
| [24] | Wang B, Wang YK, Yuan X, et al. Comparative transcriptomic analysis provides key genetic resources in clove basil (Ocimum gratissimum) under cadmium stress [J]. Front Genet, 2023, 14: 1224140. |
| [25] | Wang B, Wang G, Wang YK, et al. A cold-inducible MYB like transcription factor, CsHHO2, positively regulates chilling tolerance of cucumber fruit by enhancing CsGR-RBP3 expression [J]. Postharvest Biol Technol, 2024, 218: 113172. |
| [26] | Dong YM, Zhang WY, Li JR, et al. The transcription factor LaMYC4 from lavender regulates volatile Terpenoid biosynthesis [J]. BMC Plant Biol, 2022, 22(1): 289. |
| [27] | Vogt T. Phenylpropanoid biosynthesis [J]. Mol Plant, 2010, 3(1): 2-20. |
| [28] | Lashley A, Miller R, Provenzano S, et al. Functional diversification and structural origins of plant natural product methyltransferases [J]. Molecules, 2022, 28(1): 43. |
| [29] | Deem AK, Bultema RL, Crowell DN. Prenylcysteine methylesterase in Arabidopsis thaliana [J]. Gene, 2006, 380(2): 159-166. |
| [30] | 陈峰, 李海龙, 谭银丰, 等. 采收时期、部位、干燥方式对海南罗勒精油质量的影响 [J]. 海南医学院学报, 2012, 18(3): 297-300, 304. |
| Chen F, Li HL, Tan YF, et al. Harvest period, parts of plant, and drying methods affect the essential oils extracted from Basil(Ocimum basilicum L.) by steam distillation [J]. J Hainan Med Univ, 2012, 18(3): 297-300, 304. | |
| [31] | 李雨晴, 吴楠, 罗建让. 卵叶牡丹花色苷合成相关基因bHLH的克隆与功能分析 [J]. 生物技术通报, 2024, 40(8): 174-185. |
| Li YQ, Wu N, Luo JR. Cloning and functional analysis of bHLH gene related to anthocyanin synthesis in Paeonia qiui [J]. Biotechnol Bull, 2024, 40(8): 174-185. | |
| [32] | 周嫦, 杨弘远. 水稻未受精胚囊的离体胚胎发生 [J]. Integr Plant Biol, 1981, (3):176-180,260-261. |
| Zhou C, Yang HY. In vitro embryogenesis in unfertilized embryo sacs of Oryza sativa [J]. J Integr Plant Biol, 1981, (3): 176-180,260-261. | |
| [33] | Higashiyama T, Kuroiwa H, Kawano S, et al. Guidance in vitro of the pollen tube to the naked embryo sac of to To renia fournieri [J]. Plant Cell, 1998, 10(12): 2019-2032. |
| [34] | 李林. 基于茉莉素信号介导的丹参转录因子SmbHLH59、SmMYB97和SmWRKY14的功能研究 [D]. 西安: 陕西师范大学, 2021. |
| Li L. Functional study on transcription factors SmbHLH59, SmMYB97 and SmWRKY14 of Salvia miltiorrhiza based on jasmine signal [D]. Xi'an: Shaanxi Normal University, 2021. | |
| [35] | Oh J, Park E, Song K, et al. PHYTOCHROME INTERACTING FACTOR8 inhibits phytochrome A-mediated far-red light responses in Arabidopsis [J]. Plant Cell, 2020, 32(1): 186-205. |
| [36] | Hirose S, Sakai K, Kobayashi S, et al. Eugenol transport and biosynthesis through grafting in aromatic plants of the Ocimum genus [J]. Plant Biotechnol, 2024, 41(2): 111-120. |
| [37] | Zhang C, Chen XL, Crandall-Stotler B, et al. Biosynthesis of methyl (E)-cinnamate in the liverwort Conocephalum salebrosum and evolution of cinnamic acid methyltransferase [J]. Phytochemistry, 2019, 164: 50-59. |
| [38] | Vranová E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis [J]. Annu Rev Plant Biol, 2013, 64: 665-700. |
| [39] | Ferreira SS, Antunes MS. Re-engineering plant phenylpropanoid metabolism with the aid of synthetic biosensors [J]. Front Plant Sci, 2021, 12: 701385. |
| [40] | Wei JC, Yang Y, Peng Y, et al. Biosynthesis and the transcriptional regulation of terpenoids in tea plants (Camellia sinensis) [J]. Int J Mol Sci, 2023, 24(8): 6937. |
| [1] | YANG Chao-jie, ZHANG Lan, CHEN Hong, HUANG Juan, SHI Tao-xiong, ZHU Li-wei, CHEN Qing-fu, LI Hong-you, DENG Jiao. Functional Identification of the Transcription Factor Gene FtbHLH3 in Regulating Flavonoid Biosynthesis in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2025, 41(4): 134-144. |
| [2] | WANG Bin, WANG Yu-kun, XIAO Yan-hui. Comparative Transcriptomic Analysis of Clove Basil (Ocimum gratissimum) Leaves in Response to Cadmium Stress [J]. Biotechnology Bulletin, 2025, 41(3): 255-270. |
| [3] | LI Yu-qing, WU Nan, LUO Jian-rang. Cloning and Functional Analysis of bHLH Gene Related to Anthocyanin Synthesis in Paeonia qiui [J]. Biotechnology Bulletin, 2024, 40(8): 174-185. |
| [4] | WANG Bin, YUAN Xiao, JIANG Yuan-yuan, WANG Yu-kun, XIAO Yan-hui, HE Jin-ming. Cloning of bHLH96 Gene and Its Roles in Regulating the Biosynthesis of Peppermint Terpenes [J]. Biotechnology Bulletin, 2024, 40(1): 281-293. |
| [5] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [6] | FENG Jian-ying, LI Li-qin, LU Li-ming. Genome-wide Identification and Expression Analysis of the bHLH Transcription Factor Family in Solanum tuberosum [J]. Biotechnology Bulletin, 2022, 38(2): 21-33. |
| [7] | SHAN Cao-mei, YE Lei, ZHANG Lian-hu, KUANG Wei-gang, SUN Xiao-tang, MA Jian, CUI Ru-qiang. Cloning,and Functional Analysis of Gene OsRAI1 Resistant to Hirschmanniella mucronate in Rice [J]. Biotechnology Bulletin, 2021, 37(7): 146-155. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||