








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (9): 256-264.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0122
ZHANG Chao-chao( ), HAN Kai-yuan, WANG Tong, CHEN Zhong(
), HAN Kai-yuan, WANG Tong, CHEN Zhong( )
)
Received:2025-02-06
Online:2025-09-26
Published:2025-09-24
Contact:
CHEN Zhong
E-mail:zhangcc99@bjfu.edu.cn;zhongchen@bjfu.edu.cn
ZHANG Chao-chao, HAN Kai-yuan, WANG Tong, CHEN Zhong. Cloning and Functional Analysis of PtoYABBY2 and PtoYABBY12 in Populus tomentosa[J]. Biotechnology Bulletin, 2025, 41(9): 256-264.
引物名称 Primer name | 正向引物 Forward primer (5′‒3′) | 反向引物 Reverse primer (5′‒3′) | 用途 Purpose |
|---|---|---|---|
| PtoYAB2 | ATGCATCACATAAACGAAAGGAAA | TCACAGGGAGCCATTGATCTCGTA | 扩增PCR |
| PtoYAB12 | ATGTCTCTAGACATTGCTTCTGAACG | TTATGCATCCATGCCGGTG | |
| PtoYAB2-qPCR | CTTTGGACTGAAGCTGGACA | TCACAGGGAGCCATTGAT | RT-qPCR |
| PtoYAB12-qPCR | TCCCAGTCTTCTTCCTCGGG | GTTCTTGGCTGCATTGCTGA | |
| PtACTIN | TATCTCCTCTGTCTCCGACT | GCATCATCACCTGCAAAC | |
| AtFT (AT1G65480) | CTTGGCAGGCAAACAGTGTATGCAC | GCCACTCTCCCTCTGACAATTGTAGA | |
| AtFUL (AT5G60910) | TTGCAAGATCACAACAATTCGCTTCT | GAGAGTTTGGTTCCGTCAACGACGAT | |
| AtSOC1 (AT2G45660) | AGCTGCAGAAAACGAGAAGCTCTCTG | GGGCTACTCTCTTCATCACCTCTTCC | |
| AtLFY (AT5G61850) | ACGCCGTCATTTGCTACTCT | CTTTCTCCGTCTCTGCTGCT | |
| AtFLC (AT5G10140) | CTAGCCAGATGGAGAATAATCATCATG | TTAAGGTGGCTAATTAAGTAGTGGGAG | |
| AtACTIN2 (AT3G18780) | AGTGGTCGTACAACCGGTATTGT | GATGGCATGGAGGAAGAGAGAAAC |
Table 1 Primer sequence
引物名称 Primer name | 正向引物 Forward primer (5′‒3′) | 反向引物 Reverse primer (5′‒3′) | 用途 Purpose |
|---|---|---|---|
| PtoYAB2 | ATGCATCACATAAACGAAAGGAAA | TCACAGGGAGCCATTGATCTCGTA | 扩增PCR |
| PtoYAB12 | ATGTCTCTAGACATTGCTTCTGAACG | TTATGCATCCATGCCGGTG | |
| PtoYAB2-qPCR | CTTTGGACTGAAGCTGGACA | TCACAGGGAGCCATTGAT | RT-qPCR |
| PtoYAB12-qPCR | TCCCAGTCTTCTTCCTCGGG | GTTCTTGGCTGCATTGCTGA | |
| PtACTIN | TATCTCCTCTGTCTCCGACT | GCATCATCACCTGCAAAC | |
| AtFT (AT1G65480) | CTTGGCAGGCAAACAGTGTATGCAC | GCCACTCTCCCTCTGACAATTGTAGA | |
| AtFUL (AT5G60910) | TTGCAAGATCACAACAATTCGCTTCT | GAGAGTTTGGTTCCGTCAACGACGAT | |
| AtSOC1 (AT2G45660) | AGCTGCAGAAAACGAGAAGCTCTCTG | GGGCTACTCTCTTCATCACCTCTTCC | |
| AtLFY (AT5G61850) | ACGCCGTCATTTGCTACTCT | CTTTCTCCGTCTCTGCTGCT | |
| AtFLC (AT5G10140) | CTAGCCAGATGGAGAATAATCATCATG | TTAAGGTGGCTAATTAAGTAGTGGGAG | |
| AtACTIN2 (AT3G18780) | AGTGGTCGTACAACCGGTATTGT | GATGGCATGGAGGAAGAGAGAAAC |
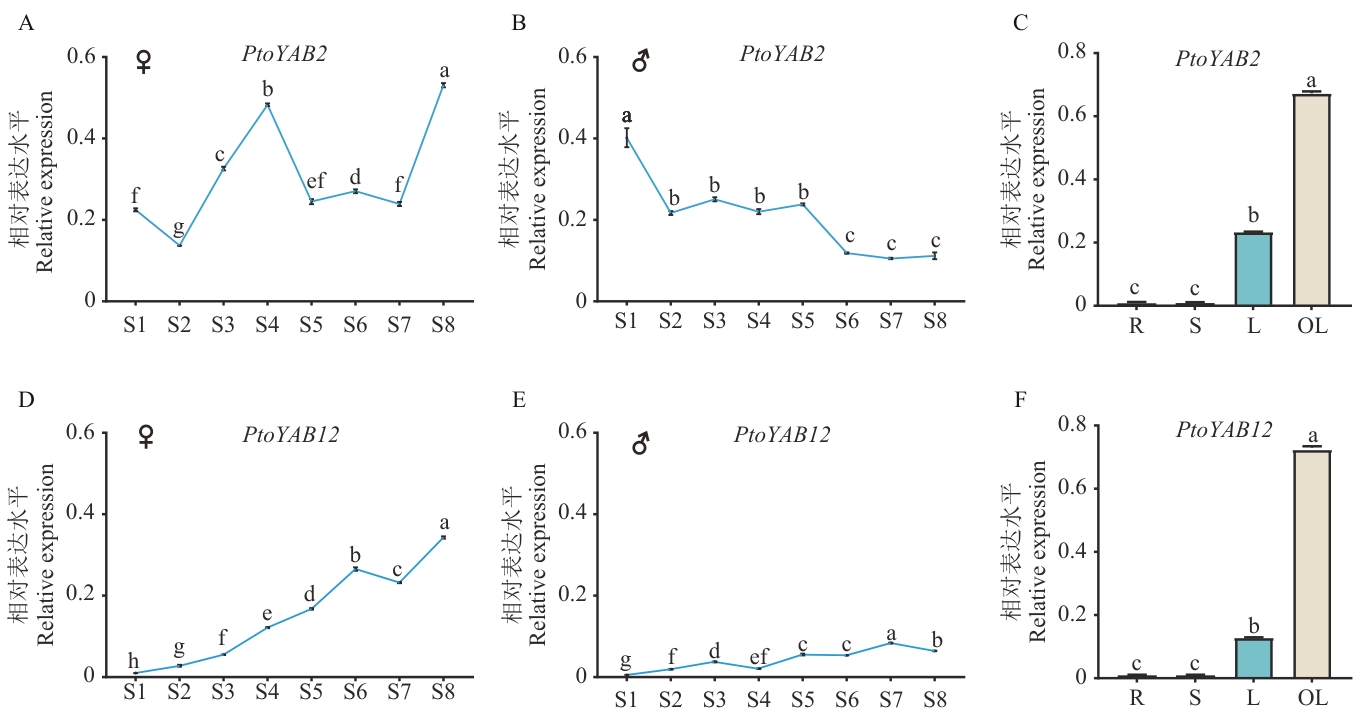
Fig. 5 Expressions of PtoYAB2 and PtoYAB12 in male and female flower buds and different tissues of P. tomentosaS1‒S8 refer to the flower buds at the eight key developmental stages of flower bud development (S1 flower induction stage, S2 flower primordium formation stage, S3 organogenesis stage, S4 elongation stage, S5 archespore formation stage, S6 and S7 dormancy stage, and S8 archespore genesis stage); R, S, L and OL refer to the root, stem, young leaf and mature leaf respectively. ♀ indicates female flower buds and ♂ indicates male flower buds. Different lowercase letters indicate significant differences at the 0.05 level. The same below
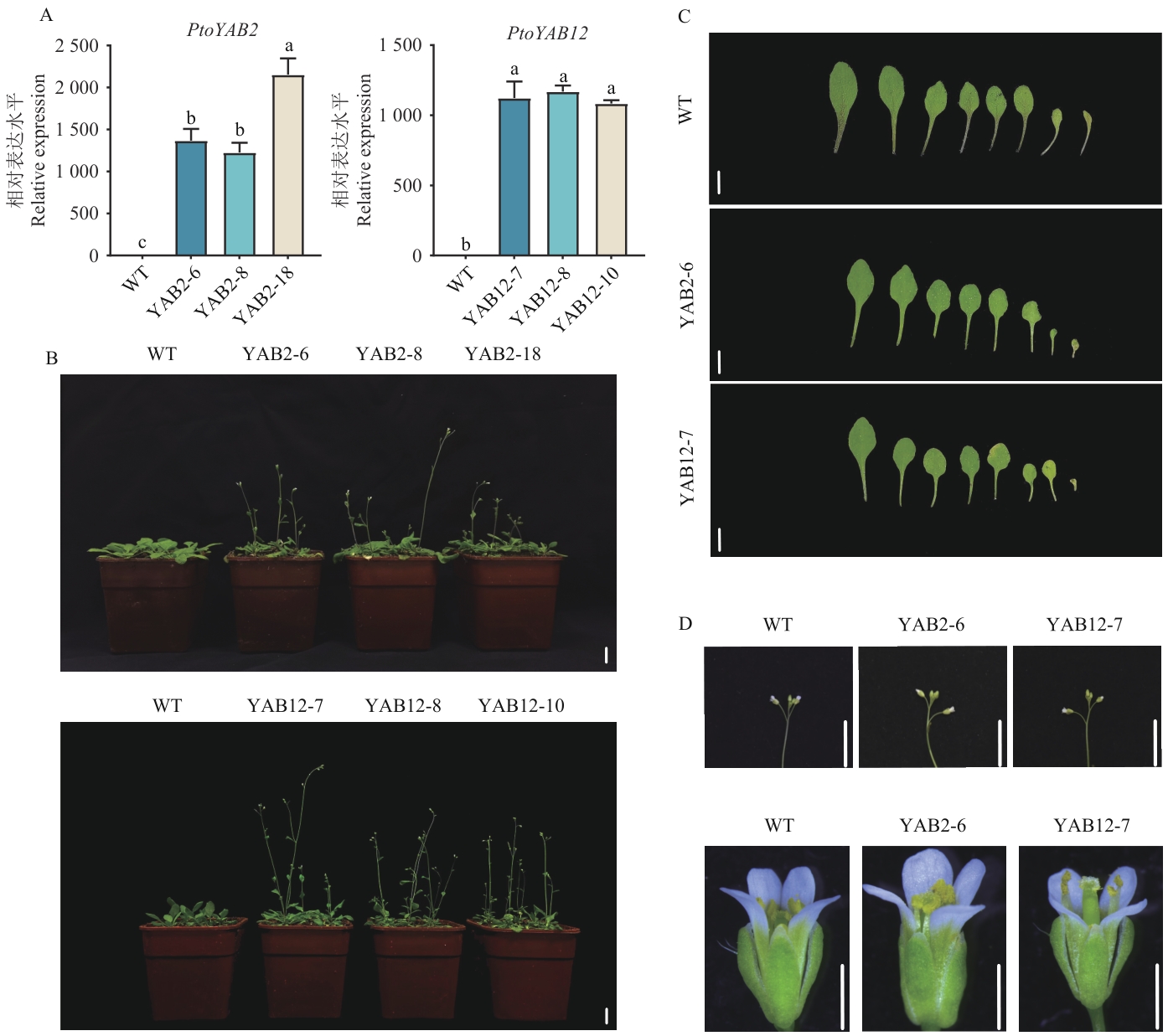
Fig. 6 Identification and phenotypic analysis of transgenic A. thalianaA: Positive identification of transgenic Arabidopsis thaliana. B: Comparison of flowering time between transgenic A. thaliana and wild type. C: Comparison of leaf phenotypes between transgenic A. thaliana and wild type. D: Comparison of indeterminate inflorescences and floral organs between transgenic A. thaliana and wild type. YAB2-6, YAB2-8, and YAB2-18 are PtoYAB2 transgenic A. thaliana lines, and YAB12-7, YAB12-8, and YAB12-10 are PtoYAB12 transgenic A. thaliana lines, the same below. The scale of B and C is 1 cm, the scale of D from top to bottom is 1 cm and 1mm
株系 Line | 光照条件 Light condition | 植株数 Number of plants | 开花时间 Time of anthesis (d) |
|---|---|---|---|
| WT (Col) | LD | 10 | 20.33±0.36 |
| YAB2-6 | LD | 11 | 16.00±0.25** |
| YAB2-8 | LD | 12 | 16.50±0.36** |
| YAB2-18 | LD | 10 | 16.00±0.37** |
| YAB12-7 | LD | 12 | 16.16±0.37** |
| YAB12-8 | LD | 12 | 16.00±0.30** |
| YAB12-10 | LD | 10 | 15.75±0.25** |
Table 2 Bolting flowering time of wild-type and transgenic A. thaliana
株系 Line | 光照条件 Light condition | 植株数 Number of plants | 开花时间 Time of anthesis (d) |
|---|---|---|---|
| WT (Col) | LD | 10 | 20.33±0.36 |
| YAB2-6 | LD | 11 | 16.00±0.25** |
| YAB2-8 | LD | 12 | 16.50±0.36** |
| YAB2-18 | LD | 10 | 16.00±0.37** |
| YAB12-7 | LD | 12 | 16.16±0.37** |
| YAB12-8 | LD | 12 | 16.00±0.30** |
| YAB12-10 | LD | 10 | 15.75±0.25** |
| [1] | Finet C, Floyd SK, Conway SJ, et al. Evolution of the YABBY gene family in seed plants [J]. Evol Dev, 2016, 18(2): 116-126. |
| [2] | Han HQ, Liu Y, Jiang MM, et al. Identification and expression analysis of YABBY family genes associated with fruit shape in tomato (Solanum lycopersicum L.) [J]. Genet Mol Res, 2015, 14(2): 7079-7091. |
| [3] | Zhao SP, Lu D, Yu TF, et al. Genome-wide analysis of the YABBY family in soybean and functional identification of GmYABBY10 involvement in high salt and drought stresses [J]. Plant Physiol Biochem, 2017, 119: 132-146. |
| [4] | Bonaccorso O, Lee JE, Puah L, et al. FILAMENTOUS FLOWER controls lateral organ development by acting as both an activator and a repressor [J]. BMC Plant Biol, 2012, 12: 176. |
| [5] | Bowman JL, Smyth DR. CRABS CLAW a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains [J]. Development, 1999, 126(11): 2387-2396. |
| [6] | Zhang XL, Yang ZP, Zhang J, et al. Ectopic expression of BraYAB1-702, a member of YABBY gene family in Chinese cabbage, causes leaf curling, inhibition of development of shoot apical meristem and flowering stage delaying in Arabidopsis thaliana [J]. Int J Mol Sci, 2013, 14(7): 14872-14891. |
| [7] | Zhang X, Ding L, Song AP, et al. Dwarf and robust plant regulates plant height via modulating gibberellin biosynthesis in Chrysanthemum [J]. Plant Physiol, 2022, 190(4): 2484-2500. |
| [8] | Huang ZJ, Van Houten J, Gonzalez G, et al. Genome-wide identification, phylogeny and expression analysis of SUN OFP and YABBY gene family in tomato [J]. Mol Genet Genomics, 2013, 288(3/4): 111-129. |
| [9] | Yang ZE, Gong Q, Wang LL, et al. Genome-wide study of YABBY genes in upland cotton and their expression patterns under different stresses [J]. Front Genet, 2018, 9: 33. |
| [10] | Han KY, Lai M, Zhao TY, et al. Plant YABBY transcription factors: a review of gene expression, biological functions, and prospects [J]. Crit Rev Biotechnol, 2025, 45(1): 214-235. |
| [11] | Yang H, Shi GX, Li X, et al. Overexpression of a soybean YABBY gene, GmFILa, causes leaf curling in Arabidopsis thaliana [J]. BMC Plant Biol, 2019, 19(1): 234. |
| [12] | Hou HL, Lin Y, Hou XL. Ectopic expression of a pak-choi YABBY gene, BcYAB3, causes leaf curvature and flowering stage delay in Arabidopsis thaliana [J]. Genes, 2020, 11(4): 370. |
| [13] | 郭效诚. 蜡梅YABBY家族CpYABBY5-1与CpCRC功能研究 [D]. 重庆: 西南大学, 2023. |
| Guo XC. Study on CpYABBY5-1 and CpCRC function of Chimonanthus praecox YABBY family [D]. Chongqing: Southwest University, 2023. | |
| [14] | Sun XD, Guan YL, Hu XY. Isolation and characterization of IaYABBY2 gene from Incarvillea arguta [J]. Plant Mol Biol Report, 2014, 32(6): 1219-1227. |
| [15] | Liu SN, Li XF, Yang H, et al. Ectopic expression of BoYAB1, a member of YABBY gene family in Bambusa oldhamii, causes leaf curling and late flowering in Arabidopsis thaliana [J]. J Hortic Sci Biotechnol, 2020, 95(2): 169-174. |
| [16] | Ionescu IA, Møller BL, Sánchez-Pérez R. Chemical control of flowering time [J]. J Exp Bot, 2017, 68(3): 369-382. |
| [17] | Gaudinier A, Blackman BK. Evolutionary processes from the perspective of flowering time diversity [J]. New Phytol, 2020, 225(5): 1883-1898. |
| [18] | Zhang TP, Li CY, Li DX, et al. Roles of YABBY transcription factors in the modulation of morphogenesis, development, and phytohormone and stress responses in plants [J]. J Plant Res, 2020, 133(6): 751-763. |
| [19] | Li J, Chen TT, Gao K, et al. Unravelling the novel sex determination genotype with ‘ZY’ and a distinctive 2.15-2.95 Mb inversion among poplar species through haplotype-resolved genome assembly and comparative genomics analysis [J]. Mol Ecol Resour, 2024, 24(7): e14002. |
| [20] | Chen Z, Rao P, Yang XY, et al. A global view of transcriptome dynamics during male floral bud development in Populus tomentosa [J]. Sci Rep, 2018, 8(1): 722. |
| [21] | 郝颖颖, 周明珠, 韩开元, 等. 毛白杨PtYABBY基因家族3个成员的克隆与表达分析 [J]. 北京林业大学学报, 2022, 44(1): 48-57. |
| Hao YY, Zhou MZ, Han KY, et al. Cloning and expression analysis of three members of PtYABBYs in Populus tomentosa [J]. J Beijing For Univ, 2022, 44(1): 48-57. | |
| [22] | Xiang J, Liu RQ, Li TM, et al. Isolation and characterization of two VpYABBY genes from wild Chinese Vitis pseudoreticulata [J]. Protoplasma, 2013, 250(6): 1315-1325. |
| [23] | Jang S, Hur J, Kim SJ, et al. Ectopic expression of OsYAB1 causes extra stamens and carpels in rice [J]. Plant Mol Biol, 2004, 56(1): 133-143. |
| [24] | van der Knaap E, Chakrabarti M, Chu YH, et al. What lies beyond the eye: the molecular mechanisms regulating tomato fruit weight and shape [J]. Front Plant Sci, 2014, 5: 227. |
| [25] | Shi TT, Zhou L, Ye YF, et al. Characterization of YABBY transcription factors in Osmanthus fragrans and functional analysis of OfYABBY12 in floral scent formation and leaf morphology [J]. BMC Plant Biol, 2024, 24(1): 589. |
| [26] | Helliwell CA, Wood CC, Robertson M, et al. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex [J]. Plant J, 2006, 46(2): 183-192. |
| [27] | Shen LS, Zhang Y, Sawettalake N. A Molecular switch for FLOWERING LOCUS C activation determines flowering time in Arabidopsis [J]. Plant Cell, 2022, 34(2): 818-833. |
| [28] | Whittaker C, Dean C. The FLC locus: a platform for discoveries in epigenetics and adaptation [J]. Annu Rev Cell Dev Biol, 2017, 33: 555-575. |
| [29] | Xie YR, Zhou Q, Zhao YP, et al. FHY3 and FAR1 integrate light signals with the miR156-SPL module-mediated aging pathway to regulate Arabidopsis flowering [J]. Mol Plant, 2020, 13(3): 483-498. |
| [1] | SHI Fa-chao, JIANG Yong-hua, LIU Hai-lun, WEN Ying-jie, YAN Qian. Cloning and Functional Analysis of LcTFL1 Gene in Litchi chinensis Sonn. [J]. Biotechnology Bulletin, 2025, 41(9): 159-167. |
| [2] | LIU Jia-li, SONG Jing-rong, ZHAO Wen-yu, ZHANG Xin-yuan, ZHAO Zi-yang, CAO Yi-bo, ZHANG Ling-yun. Identification of the R2R3-MYB Gene and Expression Analysis of Flavonoid Regulatory Genes in Blueberry [J]. Biotechnology Bulletin, 2025, 41(9): 124-138. |
| [3] | LI Yu-zhen, LI Meng-dan, ZHANG Wei, PENG Ting. Functional Study of RmEXPB2 Genein Rosa multiflora Based on the Identification of the Expansin Gene Family in Rosa sp. [J]. Biotechnology Bulletin, 2025, 41(9): 182-194. |
| [4] | HUANG Shi-yu, TIAN Shan-shan, YANG Tian-wei, GAO Man-rong, ZHANG Shang-wen. Genome-wide Identification and Expression Pattern Analysis of WRI1 Gene Family in Erythropalum scandens [J]. Biotechnology Bulletin, 2025, 41(8): 242-254. |
| [5] | KANG Qin, WANG Xia, SHEN Ming-yang, XU Jing-tian, CHEN Shi-lan, LIAO Ping-yang, XU Wen-zhi, WU Wei, XU Dong-bei. Cloning and Expression Analysis of the UV-B Receptor Gene McUVR8 in Mentha canadensis L. [J]. Biotechnology Bulletin, 2025, 41(8): 255-266. |
| [6] | WANG Fang, QIAO Shuai, SONG Wei, CUI Peng-juan, LIAO An-zhong, TAN Wen-fang, YANG Song-tao. Genome-wide Identification of the IbNRT2 Gene Family and Its Expression in Sweet Potato [J]. Biotechnology Bulletin, 2025, 41(7): 193-204. |
| [7] | XU Hui-zhen, SHANTWANA Ghimire, RAJU Kharel, YUE Yun, SI Huai-jun, TANG Xun. Analysis of the Potato SUMO E3 Ligase Gene Family and Cloning and Expression Pattern of StSIZ1 [J]. Biotechnology Bulletin, 2025, 41(6): 119-129. |
| [8] | PENG Shao-zhi, WANG Deng-ke, ZHANG Xiang, DAI Xiong-ze, XU Hao, ZOU Xue-xiao. Cloning, Expression Characteristics and Functional Verification of the Pepper CaFD1 Gene [J]. Biotechnology Bulletin, 2025, 41(5): 153-164. |
| [9] | WANG Tian-xi, YANG Bing-song, PAN Rong-jun, GAI Wen-xian, LIANG Mei-xia. Identification of the Apple PLATZ Gene Family and Functional Study of the MdPLATZ9 Gene [J]. Biotechnology Bulletin, 2025, 41(4): 176-187. |
| [10] | SONG Jia-yi, SU Yun-li, ZHENG Xing-yan, XIA Wen-nian, YANG Dong-mei, HU Hui-zhen. Identification of the Snapdragon Expansin Gene Family and Screening of Its Genes Related to Resistance to Sclerotinia sclerotiorum [J]. Biotechnology Bulletin, 2025, 41(4): 227-242. |
| [11] | SONG Shu-yi, JIANG Kai-xiu, LIU Huan-yan, HUANG Ya-cheng, LIU Lin-ya. Identification of the TCP Gene Family in Actinidia chinensis var. Hongyang and Their Expression Analysis in Fruit [J]. Biotechnology Bulletin, 2025, 41(3): 190-201. |
| [12] | KUANG Jian-hua, CHENG Zhi-peng, ZHAO Yong-jing, YANG Jie, CHEN Run-qiao, CHEN Long-qing, HU Hui-zhen. Expression Analysis of the GH3 Gene Family in Nelumbo nucifera underHormonal and Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(2): 221-233. |
| [13] | HUANG Ying, YU Wen-jing, LIU Xue-feng, DIAO Gui-ping. Bioinformatics and Expression Pattern Analysis of Glutathione S-transferase in Populus davidiana × P. bolleana [J]. Biotechnology Bulletin, 2025, 41(2): 248-256. |
| [14] | YANG Yong, YUAN Guo-mei, KANG Xiao-xiao, LIU Ya-ming, WANG Dong-sheng, ZHANG Hai-e. Identification and Expression Analysis of Members of the SWEET Gene Family in Chinese Chestnut [J]. Biotechnology Bulletin, 2025, 41(2): 257-269. |
| [15] | DU Pin-ting, WU Guo-jiang, WANG Zhen-guo, LI Yan, ZHOU Wei, ZHOU Ya-xing. Identification and Expression Analysis of CPP Gene Family in Sorghum [J]. Biotechnology Bulletin, 2025, 41(1): 132-142. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||