








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (1): 13-30.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0643
Previous Articles Next Articles
FEI Si-tian1,2( ), HOU Ying-xiang1,2, LI Lan3, ZHANG Chao1,2(
), HOU Ying-xiang1,2, LI Lan3, ZHANG Chao1,2( )
)
Received:2025-06-19
Online:2026-01-26
Published:2026-02-04
Contact:
ZHANG Chao
E-mail:feisitian@xnu.edu.cn;ricezhangchao@xnu.edu.cn
FEI Si-tian, HOU Ying-xiang, LI Lan, ZHANG Chao. Biological Functions and Regulatory Network of SLR1, a Negative Regulator of Gibberellin Signaling in Rice[J]. Biotechnology Bulletin, 2026, 42(1): 13-30.
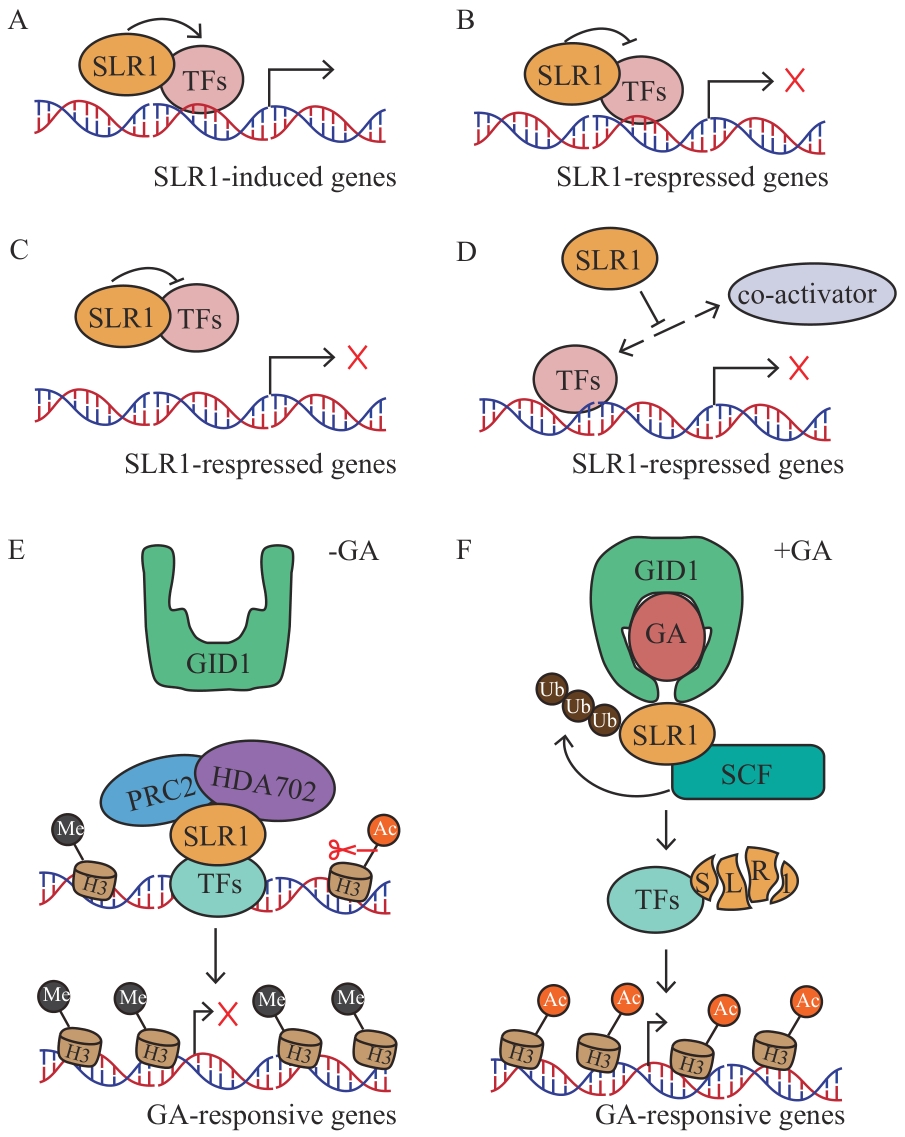
Fig. 2 Schematic diagram of the regulatory mechanism of downstream gene expression mediated by SLR1A: SLR1 binds to TFs and activates their transcriptional activity, thereby positively regulating the expressions of downstream genes. B: SLR1 binds to TFs and inhibits their transcriptional activity, thus negatively regulating the expressions of downstream genes. C: SLR1 binds to TFs to form a repressor complex, preventing TFs from binding to the cis-acting elements of downstream genes. D: SLR1 binds to transcriptional co-activators and blocks the binding between TFs and the latter, inhibiting the expressions of target genes. E: In the absence of GA, SLR1 forms a ternary complex with PRC2 and HDA702, maintaining high H3K27me3 levels and low H3K9ac levels on GA-responsive genes and inhibiting their expression. F: In the presence of sufficient GA, SLR1 is ubiquitinated and degraded, unable to form complexes with PRC2 and HDA702, thus relieving the inhibition of GA-responsive genes
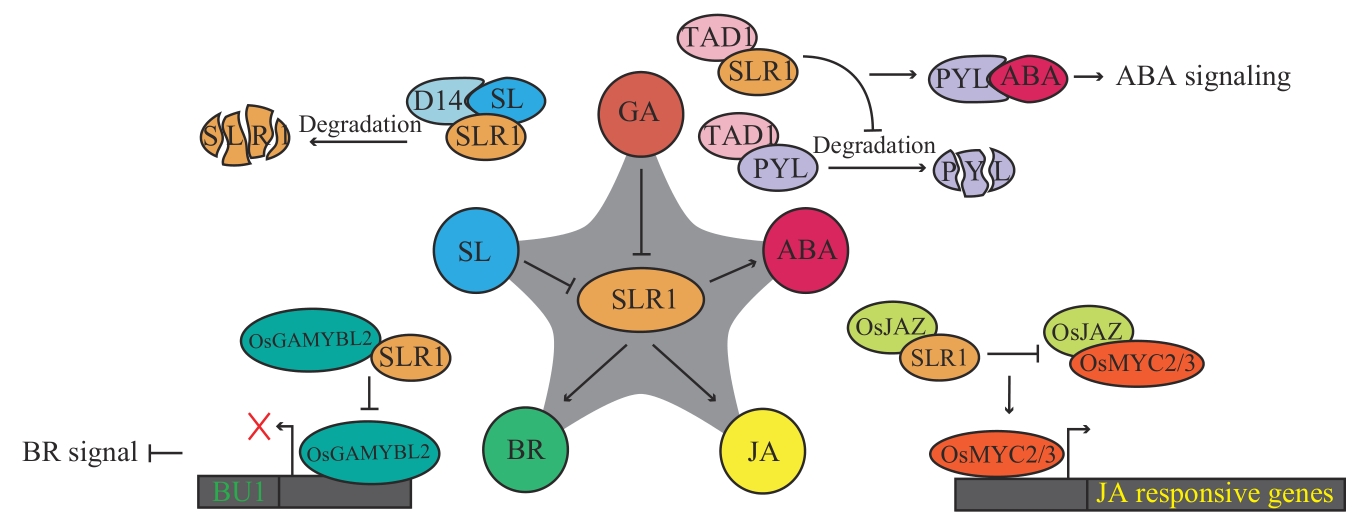
Fig. 3 SLR1 involved in other plant hormone signaling pathwaysCircles refer to plant hormones, ellipses refers to regulatory proteins in hormone pathways, split ellipses refer to protein degradation, and “→” indicates promotion, while “┫” or “×” indicates inhibition
相关基因或蛋白 Related genes or proteins | 相关通路 Related pathways | 生物学功能 Biological function | 参考文献 Reference |
|---|---|---|---|
| GID1, GID2 | GA | 介导SLR1的降解,调控GA信号途径 Mediate the degradation of SLR1 and regulate the GA signaling pathway | [ |
| D14 | GA, SL | 促进SLR1的降解,调控水稻分蘖 Promote the degradation of SLR1 and regulate rice tillering | [ |
| D53 | SL, 氮素信号 | 高氮条件下,抑制D14介导SLR1的降解,调控氮素利用 Under high-nitrogen conditions, inhibit D14-mediated degradation of SLR1 to regulate nitrogen utilization | [ |
| PRC2, HDA702 | GA | 介导染色质沉默 Mediate chromatin silencing | [ |
| EL1 | GA | 促进SLR1磷酸化,调控GA信号途径 Promote the phosphorylation of SLR1, regulate the GA signaling pathway | [ |
| OsSPY | GA | 激活SLR1的抑制活性 Activate the inhibitory activity of SLR1 | [ |
| OsYABBY4 | GA | 调控小穗发育 Regulate spikelet development | [ |
| OsWRKY36 | GA | 抑制GA信号和调控植株 Inhibit GA signaling and regulate plant height | [ |
| OsMYB91 | GA, ABA | 平衡水稻生长和非生物胁迫抗性 Balance rice growth and abiotic stress resistance | [ |
| RTD1 | GA | 负调控SLR1蛋白的转录和蛋白水平 Negatively regulate the transcription and protein level of SLR1 protein | [ |
| OsNPC6 | GA | 调控中胚轴伸长 Regulate mesocotyl elongation | [ |
| CIPK31 | GA | 抑制SLR1蛋白降解 Inhibit the degradation of SLR1 protein | [ |
| OsGAMYBL2 | GA, BR | 调控GA合成和BR信号 Regulate GA biosynthesis and BR signaling | [ |
| OsJAZ8, OsJAZ9 | JA | 调控水稻生长发育与抗逆 Regulate rice growth and development as well as stress resistance | [ |
| MYC2/3 | JA | 调控JA响应基因的表达 Regulate the expression of JA-responsive genes | [ |
| SP8, P2, M | GA, JA | 介导广谱抗病毒防御反应 Mediate broad-spectrum antiviral defense response | [ |
| TAD1 | ABA | 抑制ABA信号 Inhibit ABA signaling | [ |
| MOC1 | GA | 调控株高和分蘖 Regulate plant height and tillering | [ |
| NGR5 | GA | 提高氮素利用、促进分蘖 Enhance nitrogen utilization and promote tillering | [ |
| OsMADS23 | GA, SL | 抑制D14基因转录,促进分蘖 Suppress the transcription of the D14 gene and promote tillering | [ |
| OsIDD2 | GA | 调控细胞增殖 Regulate cell proliferation | [ |
| OsNAC29/31 | GA | 调控纤维素合成 Regulate cellulose synthesis | [ |
| OsKNAT7 | GA | 调控次生细胞壁的合成 Regulate the synthesis of secondary cell walls | [ |
| OsMYB103L | GA | 调控纤维素生物和次生细胞壁合成 Regulate cellulose and secondary cell wall biosynthesis | [ |
| OsNAC055 | 木质素合成途径 | 调控木质素合成 Regulate lignin biosynthesis | [ |
| GAMYB | GA | 调控水稻育性 Regulate rice fertility | [ |
| UDT1, TDR | GA | 提高绒毡层发育相关基因的表达,调控水稻的育性 Enhance the expressions of tapetum development-related genes and regulate rice fertility | [ |
| OsMS188 | GA | 调控孢粉素生物合成,促进花粉壁的形成 Regulate sporopollenin biosynthesis and promote pollen wall formation | [ |
| OSH1 | GA | 调控次生细胞壁合成 Regulate secondary cell wall biosynthesis | [ |
| RID1 | GA | 调控营养生长向生殖生长的转变 Regulate the transition from vegetative growth to reproductive growth | [ |
| GHD7 | GA | 调控抽穗期和植株形态 Regulate rice heading date and plant morphology | [ |
| qSH1, OSH15, SNB | GA | 调控木质素含量和落粒性 Regulate lignin biosynthesis and seed shattering | [ |
| OsNAC120 | GA, ABA | 调控干旱胁迫响应 Regulate the response to drought stress | [ |
| OsBURP3, OsSUS1 | GA | 调控干旱胁迫响应 Regulate the response to drought stress | [ |
| OsPIL13/14 | GA, 光信号 | 调控黑暗或盐胁迫下的幼苗生长 Regulate seedling growth under darkness or salt stress | [ |
| OsNF-YA3 | GA, ABA | 调控生长发育与渗透胁迫耐受性 Regulate growth and development as well as osmotic stress tolerance | [ |
| IDD10, bZIP23 | GA, 氮素信号 | 调控氮素吸收和盐碱耐受性 Regulate nitrogen absorption and saline-alkali tolerance | [ |
| OsGRF6 | GA | 调控水稻生长和耐寒性 Regulate rice growth and cold tolerance | [ |
| OsNuCYP20-2 | GA | 促进SLR1的降解,促进细胞伸长 Promote the degradation of SLR1 and cell elongation | [ |
| Sub1A | GA, ET, ABA | 促进SLR1的积累,抑制GA信号 Promote the accumulation of SLR1 and inhibit GA signaling | [ |
| GRF4 | GA, BR | 调节根系代谢和氮素利用 Regulate root metabolism and nitrogen utilization | [ |
| OsWRKY71 | GA | 调控逆境下的根系发育 Regulate root development under stress conditions | [ |
| OsSPL7, OsSPL14 | GA | 调控Xoo的抗性 Regulate resistance to Xanthomonasoryzae pv. oryzae (Xoo) | [ |
| LPA1 | GA | 对水稻纹枯病rice sheath blight(ShB)的抗性 Resistance to rice sheath blight (ShB) | [ |
Table 1 Biological functions of genes or proteins associated with SLR1
相关基因或蛋白 Related genes or proteins | 相关通路 Related pathways | 生物学功能 Biological function | 参考文献 Reference |
|---|---|---|---|
| GID1, GID2 | GA | 介导SLR1的降解,调控GA信号途径 Mediate the degradation of SLR1 and regulate the GA signaling pathway | [ |
| D14 | GA, SL | 促进SLR1的降解,调控水稻分蘖 Promote the degradation of SLR1 and regulate rice tillering | [ |
| D53 | SL, 氮素信号 | 高氮条件下,抑制D14介导SLR1的降解,调控氮素利用 Under high-nitrogen conditions, inhibit D14-mediated degradation of SLR1 to regulate nitrogen utilization | [ |
| PRC2, HDA702 | GA | 介导染色质沉默 Mediate chromatin silencing | [ |
| EL1 | GA | 促进SLR1磷酸化,调控GA信号途径 Promote the phosphorylation of SLR1, regulate the GA signaling pathway | [ |
| OsSPY | GA | 激活SLR1的抑制活性 Activate the inhibitory activity of SLR1 | [ |
| OsYABBY4 | GA | 调控小穗发育 Regulate spikelet development | [ |
| OsWRKY36 | GA | 抑制GA信号和调控植株 Inhibit GA signaling and regulate plant height | [ |
| OsMYB91 | GA, ABA | 平衡水稻生长和非生物胁迫抗性 Balance rice growth and abiotic stress resistance | [ |
| RTD1 | GA | 负调控SLR1蛋白的转录和蛋白水平 Negatively regulate the transcription and protein level of SLR1 protein | [ |
| OsNPC6 | GA | 调控中胚轴伸长 Regulate mesocotyl elongation | [ |
| CIPK31 | GA | 抑制SLR1蛋白降解 Inhibit the degradation of SLR1 protein | [ |
| OsGAMYBL2 | GA, BR | 调控GA合成和BR信号 Regulate GA biosynthesis and BR signaling | [ |
| OsJAZ8, OsJAZ9 | JA | 调控水稻生长发育与抗逆 Regulate rice growth and development as well as stress resistance | [ |
| MYC2/3 | JA | 调控JA响应基因的表达 Regulate the expression of JA-responsive genes | [ |
| SP8, P2, M | GA, JA | 介导广谱抗病毒防御反应 Mediate broad-spectrum antiviral defense response | [ |
| TAD1 | ABA | 抑制ABA信号 Inhibit ABA signaling | [ |
| MOC1 | GA | 调控株高和分蘖 Regulate plant height and tillering | [ |
| NGR5 | GA | 提高氮素利用、促进分蘖 Enhance nitrogen utilization and promote tillering | [ |
| OsMADS23 | GA, SL | 抑制D14基因转录,促进分蘖 Suppress the transcription of the D14 gene and promote tillering | [ |
| OsIDD2 | GA | 调控细胞增殖 Regulate cell proliferation | [ |
| OsNAC29/31 | GA | 调控纤维素合成 Regulate cellulose synthesis | [ |
| OsKNAT7 | GA | 调控次生细胞壁的合成 Regulate the synthesis of secondary cell walls | [ |
| OsMYB103L | GA | 调控纤维素生物和次生细胞壁合成 Regulate cellulose and secondary cell wall biosynthesis | [ |
| OsNAC055 | 木质素合成途径 | 调控木质素合成 Regulate lignin biosynthesis | [ |
| GAMYB | GA | 调控水稻育性 Regulate rice fertility | [ |
| UDT1, TDR | GA | 提高绒毡层发育相关基因的表达,调控水稻的育性 Enhance the expressions of tapetum development-related genes and regulate rice fertility | [ |
| OsMS188 | GA | 调控孢粉素生物合成,促进花粉壁的形成 Regulate sporopollenin biosynthesis and promote pollen wall formation | [ |
| OSH1 | GA | 调控次生细胞壁合成 Regulate secondary cell wall biosynthesis | [ |
| RID1 | GA | 调控营养生长向生殖生长的转变 Regulate the transition from vegetative growth to reproductive growth | [ |
| GHD7 | GA | 调控抽穗期和植株形态 Regulate rice heading date and plant morphology | [ |
| qSH1, OSH15, SNB | GA | 调控木质素含量和落粒性 Regulate lignin biosynthesis and seed shattering | [ |
| OsNAC120 | GA, ABA | 调控干旱胁迫响应 Regulate the response to drought stress | [ |
| OsBURP3, OsSUS1 | GA | 调控干旱胁迫响应 Regulate the response to drought stress | [ |
| OsPIL13/14 | GA, 光信号 | 调控黑暗或盐胁迫下的幼苗生长 Regulate seedling growth under darkness or salt stress | [ |
| OsNF-YA3 | GA, ABA | 调控生长发育与渗透胁迫耐受性 Regulate growth and development as well as osmotic stress tolerance | [ |
| IDD10, bZIP23 | GA, 氮素信号 | 调控氮素吸收和盐碱耐受性 Regulate nitrogen absorption and saline-alkali tolerance | [ |
| OsGRF6 | GA | 调控水稻生长和耐寒性 Regulate rice growth and cold tolerance | [ |
| OsNuCYP20-2 | GA | 促进SLR1的降解,促进细胞伸长 Promote the degradation of SLR1 and cell elongation | [ |
| Sub1A | GA, ET, ABA | 促进SLR1的积累,抑制GA信号 Promote the accumulation of SLR1 and inhibit GA signaling | [ |
| GRF4 | GA, BR | 调节根系代谢和氮素利用 Regulate root metabolism and nitrogen utilization | [ |
| OsWRKY71 | GA | 调控逆境下的根系发育 Regulate root development under stress conditions | [ |
| OsSPL7, OsSPL14 | GA | 调控Xoo的抗性 Regulate resistance to Xanthomonasoryzae pv. oryzae (Xoo) | [ |
| LPA1 | GA | 对水稻纹枯病rice sheath blight(ShB)的抗性 Resistance to rice sheath blight (ShB) | [ |
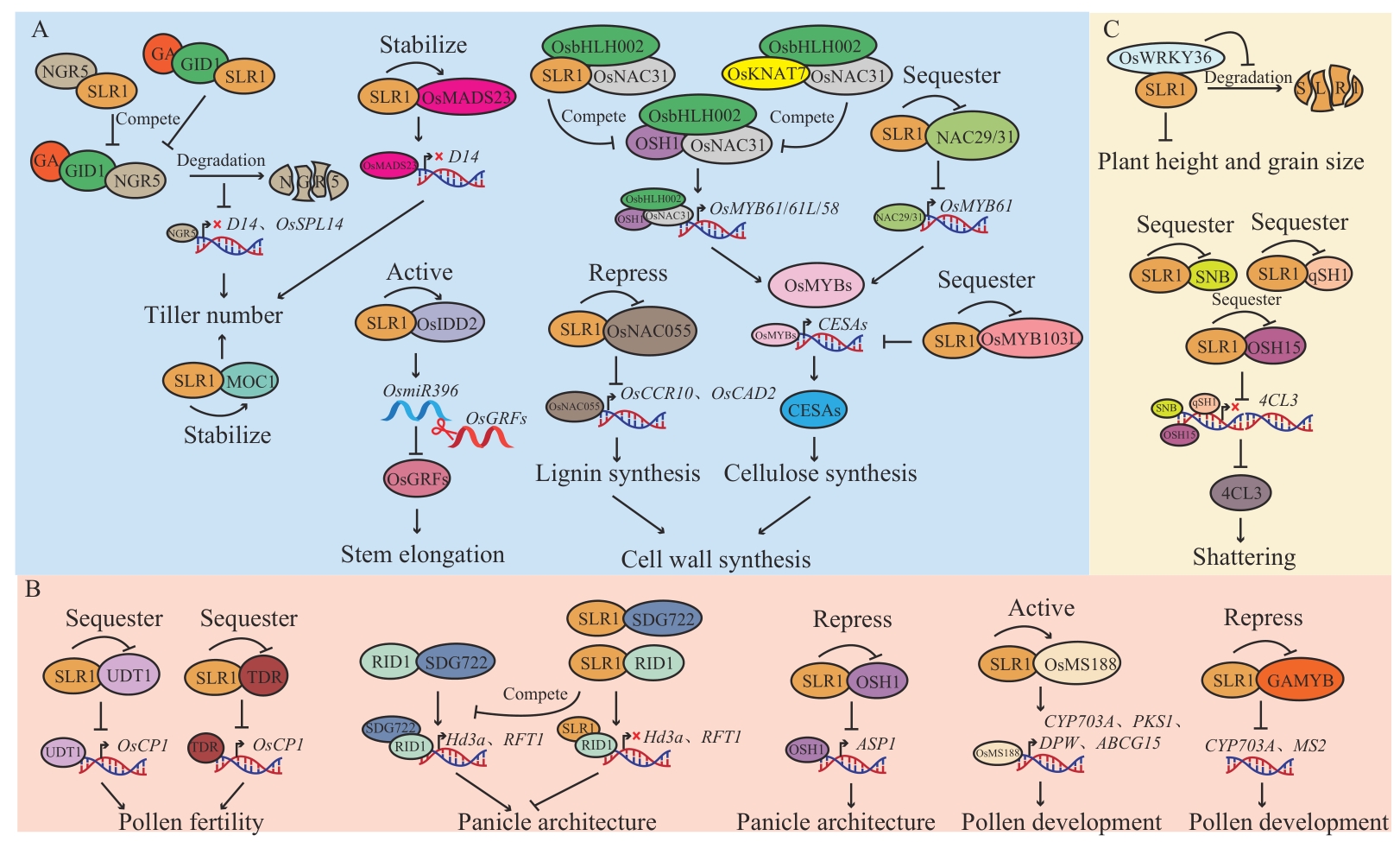
Fig. 4 SLR1 regulates rice growth and developmentA: SLR1 is involved in regulating the vegetative growth of rice; B: SLR1 is involved in regulating rice flower development; C: SLR1 is involved in regulating rice seed development. Ellipses rindicate regulatory proteins in rice growth and development pathways, split ellipses indicate protein degradation, “→” indicates promotion, and “┫” or “×” indicates inhibition
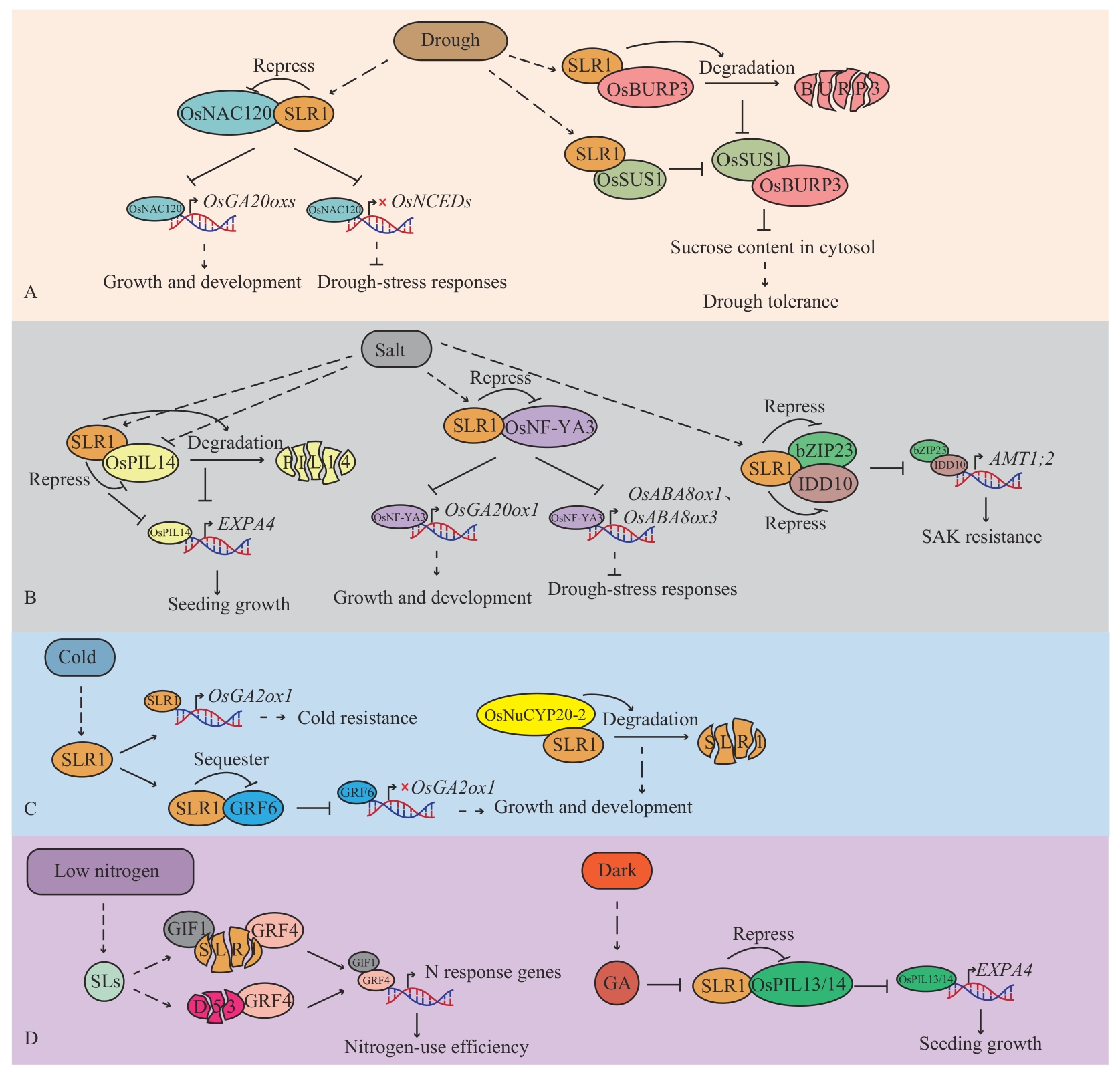
Fig. 5 SLR1 regulates abiotic stressA: SLR1 regulates drought stress; B: SLR1 regulates salt stress; C: SLR1 regulates cold stress; D: SLR1 regulates low-nitrogen stress. Rounded rectangles indicate abiotic stresses, Ellipses indicate regulatory proteins in the rice abiotic stress response pathway, split ellipses indicate protein degradation, “→” indicates promotion, “┫” or “×” indicates inhibition, and dashed arrows indicate indirect promotion
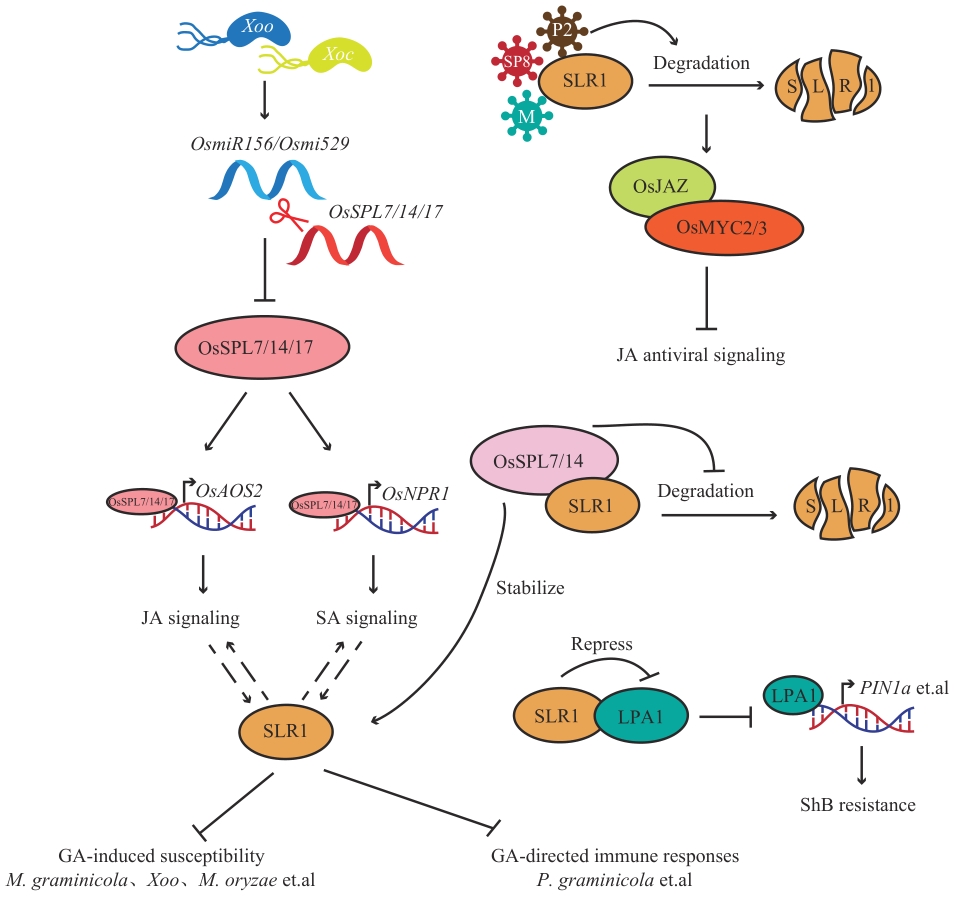
Fig. 6 SLR1 regulates biological stressEllipses indicate regulatory proteins in the rice biotic stress response pathway, split ellipses indicate protein degradation, “→” indicates promotion, “┫” indicates inhibition, and dashed arrows indicate indirect promotion
| [1] | Hedden P, Proebsting WM. Genetic analysis of gibberellin biosynthesis [J]. Plant Physiol, 1999, 119(2): 365-370. |
| [2] | Olszewski N, Sun TP, Gubler F. Gibberellin signaling: biosynthesis, catabolism, and response pathways [J]. Plant Cell, 2002, 14(): S61-S80. |
| [3] | Richards DE, King KE, Ait-Ali T, et al. How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling [J]. Annu Rev Plant Physiol Plant Mol Biol, 2001, 52: 67-88. |
| [4] | Khush GS. Green revolution: preparing for the 21st century [J]. Genome, 1999, 42(4): 646-655. |
| [5] | Pingali PL. Green revolution: impacts, limits, and the path ahead [J]. Proc Natl Acad Sci USA, 2012, 109(31): 12302-12308. |
| [6] | Monna L, Kitazawa N, Yoshino R, et al. Positional cloning of rice semidwarfing gene, sd-1: rice "green revolution gene" encodes a mutant enzyme involved in gibberellin synthesis [J]. DNA Res, 2002, 9(1): 11-17. |
| [7] | Spielmeyer W, Ellis MH, Chandler PM. Semidwarf (sd-1), "green revolution" rice, contains a defective gibberellin 20-oxidase gene [J]. Proc Natl Acad Sci USA, 2002, 99(13): 9043-9048. |
| [8] | Hedden P. The genes of the green revolution [J]. Trends Genet, 2003, 19(1): 5-9. |
| [9] | Sun TP, Gubler F. Molecular mechanism of gibberellin signaling in plants [J]. Annu Rev Plant Biol, 2004, 55: 197-223. |
| [10] | Pysh LD, Wysocka-Diller JW, Camilleri C, et al. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes [J]. Plant J, 1999, 18(1): 111-119. |
| [11] | Ogawa M, Kusano T, Katsumi M, et al. Rice gibberellin-insensitive gene homolog, OsGAI, encodes a nuclear-localized protein capable of gene activation at transcriptional level [J]. Gene, 2000, 245(1): 21-29. |
| [12] | Fu XD, Richards DE, Ait-Ali T, et al. Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor [J]. Plant Cell, 2002, 14(12): 3191-3200. |
| [13] | Jiang CF, Fu XD. GA action: turning on de-DELLA repressing signaling [J]. Curr Opin Plant Biol, 2007, 10(5): 461-465. |
| [14] | Ikeda A, Ueguchi-Tanaka M, Sonoda Y, et al. Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8 [J]. Plant Cell, 2001, 13(5): 999-1010. |
| [15] | Willige BC, Ghosh S, Nill C, et al. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis [J]. Plant Cell, 2007, 19(4): 1209-1220. |
| [16] | Ueguchi-Tanaka M, Nakajima M, Katoh E, et al. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin [J]. Plant Cell, 2007, 19(7): 2140-2155. |
| [17] | Van De Velde K, Ruelens P, Geuten K, et al. Exploiting DELLA signaling in cereals [J]. Trends Plant Sci, 2017, 22(10): 880-893. |
| [18] | Li MZ, An FY, Li WY, et al. DELLA proteins interact with FLC to repress flowering transition [J]. J Integr Plant Biol, 2016, 58(7): 642-655. |
| [19] | Gomi K, Sasaki A, Itoh H, et al. GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice [J]. Plant J, 2004, 37(4): 626-634. |
| [20] | Itoh H, Ueguchi-Tanaka M, Sato Y, et al. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei [J]. Plant Cell, 2002, 14(1): 57-70. |
| [21] | 白云赫, 朱旭东, 樊秀彩, 等. 植物DELLA蛋白及其应答赤霉素信号调控植物生长发育的研究进展 [J]. 分子植物育种, 2019, 17(8): 2509-2516. |
| Bai YH, Zhu XD, Fan XC, et al. Reseach progress of plant DELLA proteins and its response to gibberellin signal regulating plant growth and development [J]. Mol Plant Breed, 2019, 17(8): 2509-2516. | |
| [22] | Hirano K, Asano K, Tsuji H, et al. Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice [J]. Plant Cell, 2010, 22(8): 2680-2696. |
| [23] | Hellens RP, Allan AC, Friel EN, et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants [J]. Plant Methods, 2005, 1: 13. |
| [24] | Hauvermale AL, Ariizumi T, Steber CM. Gibberellin signaling: a theme and variations on DELLA repression [J]. Plant Physiol, 2012, 160(1): 83-92. |
| [25] | Ueguchi-Tanaka M. Gibberellin metabolism and signaling [J]. Biosci Biotechnol Biochem, 2023, 87(10): 1093-1101. |
| [26] | Phokas A, Coates JC. Evolution of DELLA function and signaling in land plants [J]. Evol Dev, 2021, 23(3): 137-154. |
| [27] | Yano K, Aya K, Hirano K, et al. Comprehensive gene expression analysis of rice aleurone cells: probing the existence of an alternative gibberellin receptor [J]. Plant Physiol, 2015, 167(2): 531-544. |
| [28] | Sun HW, Guo XL, Zhu XL, et al. Strigolactone and gibberellin signaling coordinately regulate metabolic adaptations to changes in nitrogen availability in rice [J]. Mol Plant, 2023, 16(3): 588-598. |
| [29] | Zhao Y, Zhou DX. Epigenomic modification and epigenetic regulation in rice [J]. J Genet Genom, 2012, 39(7): 307-315. |
| [30] | Chen XS, Zhou DX. Rice epigenomics and epigenetics: challenges and opportunities [J]. Curr Opin Plant Biol, 2013, 16(2): 164-169. |
| [31] | Li TT, Chen XS, Zhong XC, et al. Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice [J]. Plant Cell, 2013, 25(11): 4725-4736. |
| [32] | Li JJ, Li Q, Wang WT, et al. DELLA-mediated gene repression is maintained by chromatin modification in rice [J]. EMBO J, 2023, 42(21): e114220. |
| [33] | Itoh H, Matsuoka M, Steber CM. A role for the ubiquitin-26S-proteasome pathway in gibberellin signaling [J]. Trends Plant Sci, 2003, 8(10): 492-497. |
| [34] | Mashita O, Koishihara H, Fukui K, et al. Discovery and identification of 2-methoxy-1-naphthaldehyde as a novel strigolactone-signaling inhibitor [J]. J Pestic Sci, 2016, 41(3): 71-78. |
| [35] | Han SN, Liu YX, Bao A, et al. OsCSN1 regulates the growth of rice seedlings through the GA signaling pathway in blue light [J]. J Plant Physiol, 2023, 280: 153904. |
| [36] | Fernandes T, Gonçalves NM, Matiolli CC, et al. SUMOylation of rice DELLA SLR1 modulates transcriptional responses and improves yield under salt stress [J]. Planta, 2024, 260(6): 136. |
| [37] | Itoh H, Sasaki A, Ueguchi-Tanaka M, et al. Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1 [J]. Plant Cell Physiol, 2005, 46(8): 1392-1399. |
| [38] | Dai C, Xue HW. Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling [J]. EMBO J, 2010, 29(11): 1916-1927. |
| [39] | Shimada A, Ueguchi-Tanaka M, Sakamoto T, et al. The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1 and modulating brassinosteroid synthesis [J]. Plant J, 2006, 48(3): 390-402. |
| [40] | Yano K, Morinaka Y, Wang FM, et al. GWAS with principal component analysis identifies a gene comprehensively controlling rice architecture [J]. Proc Natl Acad Sci USA, 2019, 116(42): 21262-21267. |
| [41] | Yang C, Ma YM, Li JX. The rice YABBY4 gene regulates plant growth and development through modulating the gibberellin pathway [J]. J Exp Bot, 2016, 67(18): 5545-5556. |
| [42] | Lan J, Lin QB, Zhou CL, et al. Small grain and semi-dwarf 3, a WRKY transcription factor, negatively regulates plant height and grain size by stabilizing SLR1 expression in rice [J]. Plant Mol Biol, 2020, 104(4/5): 429-450. |
| [43] | Zhu N, Cheng SF, Liu XY, et al. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice [J]. Plant Sci, 2015, 236: 146-156. |
| [44] | Zhang YH, Liu K, Zhu XM, et al. Rice tocopherol deficiency 1 encodes a homogentisate phytyltransferase essential for tocopherol biosynthesis and plant development in rice [J]. Plant Cell Rep, 2018, 37(5): 775-787. |
| [45] | Yang D, Liu X, Yin XM, et al. Rice non-specific phospholipase C6 is involved in mesocotyl elongation [J]. Plant Cell Physiol, 2021, 62(6): 985-1000. |
| [46] | Chen JS, Wang ST, Jiang SQ, et al. Overexpression of Calcineurin B-like interacting protein kinase 31 promotes lodging and sheath blight resistance in rice [J]. Plants, 2024, 13(10): 1306. |
| [47] | Davière JM, Achard P. A pivotal role of DELLAs in regulating multiple hormone signals [J]. Mol Plant, 2016, 9(1): 10-20. |
| [48] | 赵春丽, 王晓, 陈家兰, 等. 植物DELLA蛋白家族研究进展 [J]. 应用与环境生物学报, 2020, 26(5): 1299-1308. |
| Zhao CL, Wang X, Chen JL, et al. Progress in research on plant DELLA family proteins [J]. Chin J Appl Environ Biol, 2020, 26(5): 1299-1308. | |
| [49] | Eckardt NA. GA perception and signal transduction: molecular interactions of the GA receptor GID1 with GA and the DELLA protein SLR1 in rice [J]. Plant Cell, 2007, 19(7): 2095-2097. |
| [50] | Asano K, Hirano K, Ueguchi-Tanaka M, et al. Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice [J]. Mol Genet Genomics, 2009, 281(2): 223-231. |
| [51] | Jung YJ, Kim JH, Lee HJ, et al. Generation and transcriptome profiling of Slr1-d7 and Slr1-d8 mutant lines with a new semi-dominant dwarf allele of SLR1 using the CRISPR/Cas9 system in rice [J]. Int J Mol Sci, 2020, 21(15): 5492. |
| [52] | Zentella R, Zhang ZL, Park M, et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis [J]. Plant Cell, 2007, 19(10): 3037-3057. |
| [53] | Tsuji H, Aya K, Ueguchi-Tanaka M, et al. GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers [J]. Plant J, 2006, 47(3): 427-444. |
| [54] | Gao J, Chen H, Yang HF, et al. A brassinosteroid responsive miRNA-target module regulates gibberellin biosynthesis and plant development [J]. New Phytol, 2018, 220(2): 488-501. |
| [55] | Hou XL, Lee LYC, Xia KF, et al. DELLAs modulate jasmonate signaling via competitive binding to JAZs [J]. Dev Cell, 2010, 19(6): 884-894. |
| [56] | Um TY, Lee HY, Lee S, et al. Jasmonate zim-domain protein 9 interacts with slender rice 1 to mediate the antagonistic interaction between jasmonic and gibberellic acid signals in rice [J]. Front Plant Sci, 2018, 9: 1866. |
| [57] | Li LL, Zhang HH, Yang ZH, et al. Independently evolved viral effectors convergently suppress DELLA protein SLR1-mediated broad-spectrum antiviral immunity in rice [J]. Nat Commun, 2022, 13(1): 6920. |
| [58] | Nakamura H, Xue YL, Miyakawa T, et al. Molecular mechanism of strigolactone perception by DWARF14 [J]. Nat Commun, 2013, 4: 2613. |
| [59] | Liao ZG, Zhang YC, Yu Q, et al. Coordination of growth and drought responses by GA-ABA signaling in rice [J]. New Phytol, 2023, 240(3): 1149-1161. |
| [60] | Liao ZG, Yu H, Duan JB, et al. SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice [J]. Nat Commun, 2019, 10(1): 2738. |
| [61] | Wu K, Wang SS, Song WZ, et al. Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice [J]. Science, 2020, 367(6478): eaaz2046. |
| [62] | Li XX, Xie ZZ, Qin T, et al. The SLR1-OsMADS23-D14 module mediates the crosstalk between strigolactone and gibberellin signaling to control rice tillering [J]. New Phytol, 2025, 246(5): 2137-2154. |
| [63] | Zhuang H, Li YF. Strigolactone and gibberellin crosstalk: the role of the SLR1-OsMADS23-D14 module in regulating rice tiller development [J]. New Phytol, 2025, 246(5): 1893-1895. |
| [64] | Lu YZ, Feng Z, Meng YL, et al. SLENDER RICE1 and Oryza sativa INDETERMINATE DOMAIN2 regulating OsmiR396 are involved in stem elongation [J]. Plant Physiol, 2020, 182(4): 2213-2227. |
| [65] | Huang DB, Wang SG, Zhang BC, et al. A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice [J]. Plant Cell, 2015, 27(6): 1681-1696. |
| [66] | Chen Y, Qi HY, Yang LJ, et al. The OsbHLH002/OsICE1-OSH1 module orchestrates secondary cell wall formation in rice [J]. Cell Rep, 2023, 42(7): 112702. |
| [67] | Ye YF, Liu BM, Zhao M, et al. CEF1/OsMYB103L is involved in GA-mediated regulation of secondary wall biosynthesis in rice [J]. Plant Mol Biol, 2015, 89(4/5): 385-401. |
| [68] | Liu YF, Wu Q, Qin ZL, et al. Transcription factor OsNAC055 regulates GA-mediated lignin biosynthesis in rice straw [J]. Plant Sci, 2022, 325: 111455. |
| [69] | Aya K, Ueguchi-Tanaka M, Kondo M, et al. Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB [J]. Plant Cell, 2009, 21(5): 1453-1472. |
| [70] | 宋欣玥. SLR1调控水稻雄蕊发育的功能研究 [D]. 上海: 上海师范大学, 2023. |
| Song XY. Study on the function of SLR1 in regulating stamen development in rice [D]. Shanghai: Shanghai Normal University, 2023. | |
| [71] | Tang JQ, Tian XJ, Mei EY, et al. WRKY53 negatively regulates rice cold tolerance at the booting stage by fine-tuning anther gibberellin levels [J]. Plant Cell, 2022, 34(11): 4495-4515. |
| [72] | Jin Y, Song XY, Chang HZ, et al. The GA-DELLA-OsMS188 module controls male reproductive development in rice [J]. New Phytol, 2022, 233(6): 2629-2642. |
| [73] | Su S, Hong J, Chen XF, et al. Gibberellins orchestrate panicle architecture mediated by DELLA-KNOX signalling in rice [J]. Plant Biotechnol J, 2021, 19(11): 2304-2318. |
| [74] | Zhang S, Deng L, Cheng R, et al. RID1 sets rice heading date by balancing its binding with SLR1 and SDG722 [J]. J Integr Plant Biol, 2022, 64(1): 149-165. |
| [75] | Liu RJ, Feng QF, Li PB, et al. GLW7.1, a strong functional allele of Ghd7, enhances grain size in rice [J]. Int J Mol Sci, 2022, 23(15): 8715. |
| [76] | Wu H, He Q, He B, et al. Gibberellin signaling regulates lignin biosynthesis to modulate rice seed shattering [J]. Plant Cell, 2023, 35(12): 4383-4404. |
| [77] | Xie ZZ, Jin L, Sun Y, et al. OsNAC120 balances plant growth and drought tolerance by integrating GA and ABA signaling in rice [J]. Plant Commun, 2024, 5(3): 100782. |
| [78] | Huang JS, Xie B, Xian FJ, et al. Gibberellin signalling mediates nucleocytoplasmic trafficking of Sucrose Synthase 1 to regulate the drought tolerance in rice [J]. Plant Biotechnol J, 2025, 23(6): 1909-1926. |
| [79] | Lyu YS, Dong XL, Niu SP, et al. An orchestrated ethylene-gibberellin signaling cascade contributes to mesocotyl elongation and emergence of rice direct seeding [J]. J Integr Plant Biol, 2024, 66(7): 1427-1439. |
| [80] | Mo WP, Tang WJ, Du YX, et al. PHYTOCHROME-INTERACTING FACTOR-LIKE14 and SLENDER RICE1 interaction controls seedling growth under salt stress [J]. Plant Physiol, 2020, 184(1): 506-517. |
| [81] | Jin XK, Zhang YF, Li XX, et al. OsNF-YA3 regulates plant growth and osmotic stress tolerance by interacting with SLR1 and SAPK9 in rice [J]. Plant J, 2023, 114(4): 914-933. |
| [82] | Li Z, Chen H, Guan QJ, et al. Gibberellic acid signaling promotes resistance to saline-alkaline stress by increasing the uptake of ammonium in rice [J]. Plant Physiol Biochem, 2024, 207: 108424. |
| [83] | Li ZT, Wang B, Zhang ZY, et al. OsGRF6 interacts with SLR1 to regulate OsGA2ox1 expression for coordinating chilling tolerance and growth in rice [J]. J Plant Physiol, 2021, 260: 153406. |
| [84] | Ge Q, Zhang YY, Xu YY, et al. Cyclophilin OsCYP20-2 with a novel variant integrates defense and cell elongation for chilling response in rice [J]. New Phytol, 2020, 225(6): 2453-2467. |
| [85] | Mittal L, Tayyeba S, Sinha AK. Finding a breather for Oryza sativa: Understanding hormone signalling pathways involved in rice plants to submergence stress [J]. Plant Cell Environ, 2022, 45(2): 279-295. |
| [86] | Fukao T, Bailey-Serres J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice [J]. Proc Natl Acad Sci USA, 2008, 105(43): 16814-16819. |
| [87] | Schmitz AJ, Folsom JJ, Jikamaru Y, et al. SUB1A-mediated submergence tolerance response in rice involves differential regulation of the brassinosteroid pathway [J]. New Phytol, 2013, 198(4): 1060-1070. |
| [88] | Li S, Tian YH, Wu K, et al. Modulating plant growth-metabolism coordination for sustainable agriculture [J]. Nature, 2018, 560(7720): 595-600. |
| [89] | Mirza Z, Gupta M. Iron reprogrammes the root system architecture by regulating OsWRKY71 in arsenic-stressed rice (Oryza sativa L.) [J]. Plant Mol Biol, 2024, 114(1): 11. |
| [90] | De Vleesschauwer D, Seifi HS, Filipe O, et al. The DELLA protein SLR1 integrates and amplifies salicylic acid- and jasmonic acid-dependent innate immunity in rice [J]. Plant Physiol, 2016, 170(3): 1831-1847. |
| [91] | Liu MM, Shi ZY, Zhang XH, et al. Inducible overexpression of Ideal Plant Architecture1 improves both yield and disease resistance in rice [J]. Nat Plants, 2019, 5(4): 389-400. |
| [92] | Sun Q, Li TY, Li DD, et al. Overexpression of Loose Plant Architecture 1 increases planting density and resistance to sheath blight disease via activation of PIN-FORMED 1a in rice [J]. Plant Biotechnol J, 2019, 17(5): 855-857. |
| [93] | Sakata T, Oda S, Tsunaga Y, et al. Reduction of gibberellin by low temperature disrupts pollen development in rice [J]. Plant Physiol, 2014, 164(4): 2011-2019. |
| [94] | Guo SQ, Chen YX, Ju YL, et al. Fine-tuning gibberellin improves rice alkali-thermal tolerance and yield [J]. Nature, 2025, 639(8053): 162-171. |
| [95] | Lu L, Chen XY, Tan QY, et al. Gibberellin-mediated sensitivity of rice roots to aluminum stress [J]. Plants, 2024, 13(4): 543. |
| [96] | Hui SG, Ke YG, Chen D, et al. Rice microRNA156/529- SQUAMOSA PROMOTER BINDING PROTEIN-LIKE7/14/17 modules regulate defenses against bacteria [J]. Plant Physiol, 2023, 192(3): 2537-2553. |
| [97] | Zhu HY, Chen H, Kantharaj V, et al. SLR1-LPA1 signal regulates sheath blight resistance and lamina joint angle in rice [J]. Plant Physiol Biochem, 2025, 222: 109689. |
| [98] | De Vleesschauwer D, Van Buyten E, Satoh K, et al. Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice [J]. Plant Physiol, 2012, 158(4): 1833-1846. |
| [99] | Zhang J, Luo T, Wang WW, et al. Silencing OsSLR1 enhances the resistance of rice to the brown planthopper Nilaparvata lugens [J]. Plant Cell Environ, 2017, 40(10): 2147-2159. |
| [100] | Yimer HZ, Nahar K, Kyndt T, et al. Gibberellin antagonizes jasmonate-induced defense against Meloidogyne graminicola in rice [J]. New Phytol, 2018, 218(2): 646-660. |
| [101] | Itoh H, Shimada A, Ueguchi-Tanaka M, et al. Overexpression of a GRAS protein lacking the DELLA domain confers altered gibberellin responses in rice [J]. Plant J, 2005, 44(4): 669-679. |
| [102] | Liu T, Gu JY, Xu CJ, et al. Overproduction of OsSLRL2 alters the development of transgenic Arabidopsis plants [J]. Biochem Biophys Res Commun, 2007, 358(4): 983-989. |
| [103] | Wang JD, Wang J, Huang LC, et al. ABA-mediated regulation of rice grain quality and seed dormancy via the NF-YB1-SLRL2-bHLH144 Module [J]. Nat Commun, 2024, 15(1): 4493. |
| [1] | YANG Yue-qin, XING Ying, ZHONG Zi-he, TIAN Wei-jun, YANG Xue-qing, WANG Jian-xu. Expression and Functional Analysis of OsMATE34 in Rice under Mercury Stress [J]. Biotechnology Bulletin, 2026, 42(1): 86-94. |
| [2] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [3] | CHENG Xue, FU Ying, CHAI Xiao-jiao, WANG Hong-yan, DENG Xin. Identification of LHC Gene Family in Setaria italica and Expression Analysis under Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(8): 102-114. |
| [4] | DENG Mei-bi, YAN Lang, ZHAN Zhi-tian, ZHU Min, HE Yu-bing. Efficient CRISPR Gene Editing in Rice Assisted by RUBY [J]. Biotechnology Bulletin, 2025, 41(8): 65-73. |
| [5] | ZHANG Xue-qiong, PAN Su-jun, LI Wei, DAI Liang-ying. Research Progress of Plant Phosphate Transporters in the Response to Stress [J]. Biotechnology Bulletin, 2025, 41(7): 28-36. |
| [6] | HOU Ying-xiang, FEI Si-tian, LI Ni, LI Lan, SONG Song-quan, WANG Wei-ping, ZHANG Chao. Research Progress in Response of Rice miRNAs to Biotic Stress [J]. Biotechnology Bulletin, 2025, 41(7): 69-80. |
| [7] | HAN Yi, HOU Chang-lin, TANG Lu, SUN Lu, XIE Xiao-dong, LIANG Chen, CHEN Xiao-qiang. Cloning and Preliminary Functional Analysis of HvERECTA Gene in Hordeum vulgare [J]. Biotechnology Bulletin, 2025, 41(7): 106-116. |
| [8] | LI Xia, ZHANG Ze-wei, LIU Ze-jun, WANG Nan, GUO Jiang-bo, XIN Cui-hua, ZHANG Tong, JIAN Lei. Cloning and Functional Study of Transcription Factor StMYB96 in Potato [J]. Biotechnology Bulletin, 2025, 41(7): 181-192. |
| [9] | GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant [J]. Biotechnology Bulletin, 2025, 41(7): 214-225. |
| [10] | WEI Yu-jia, LI Yan, KANG Yu-han, GONG Xiao-nan, DU Min, TU Lan, SHI Peng, YU Zi-han, SUN Yan, ZHANG Kun. Cloning and Expression Analysis of the CrMYB4 Gene in Carex rigescens [J]. Biotechnology Bulletin, 2025, 41(7): 248-260. |
| [11] | WU Hao, DONG Wei-feng, HE Zi-tian, LI Yan-xiao, XIE Hui, SUN Ming-zhe, SHEN Yang, SUN Xiao-li. Genome-wide Identification and Expression Analysis of the Rice BXL Gene Family [J]. Biotechnology Bulletin, 2025, 41(6): 87-98. |
| [12] | HUANG Dan, PENG Bing-yang, ZHANG Pan-pan, JIAO Yue, LYU Jia-bin. Identification of HD-Zip Gene Family in Camellia oleifera and Analysis of Its Expression under Abiotic Stress [J]. Biotechnology Bulletin, 2025, 41(6): 191-207. |
| [13] | PENG Shao-zhi, WANG Deng-ke, ZHANG Xiang, DAI Xiong-ze, XU Hao, ZOU Xue-xiao. Cloning, Expression Characteristics and Functional Verification of the Pepper CaFD1 Gene [J]. Biotechnology Bulletin, 2025, 41(5): 153-164. |
| [14] | LIU Yuan, ZHAO Ran, LU Zhen-fang, LI Rui-li. Research Progress in the Biological Metabolic Pathway and Functions of Plant Carotenoids [J]. Biotechnology Bulletin, 2025, 41(5): 23-31. |
| [15] | LIU Yuan-yuan, CHEN Xi-feng, QIAN Qian, GAO Zhen-yu. Advances in Molecular Mechanisms Regulating Panicle Development in Rice [J]. Biotechnology Bulletin, 2025, 41(5): 1-13. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||