








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (8): 176-185.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1303
Previous Articles Next Articles
CAI Guo-lei1( ), LU Xiao-kai1, LOU Shui-zhu2, YANG Hai-ying2, DU Gang1(
), LU Xiao-kai1, LOU Shui-zhu2, YANG Hai-ying2, DU Gang1( )
)
Received:2020-10-22
Online:2021-08-26
Published:2021-09-10
Contact:
DU Gang
E-mail:1733691043@qq.com;andydu60901@hotmail.com
CAI Guo-lei, LU Xiao-kai, LOU Shui-zhu, YANG Hai-ying, DU Gang. Classification and Identification of Bacillus LM Based on Whole Genome and Study on Its Antibacterial Principle[J]. Biotechnology Bulletin, 2021, 37(8): 176-185.
| 基因组组装 Genome assembly | 大小Size |
|---|---|
| 总支架数 Total number of scaffold /个 | 97 |
| 总重叠群数 Total number of contig /个 | 97 |
| 最大长度 The maximum length /bp | 365 023 |
| 最小长度 The Minimum length /bp | 300 |
| 支架大小 The scaffold size /bp | 4 000 885 |
| 重叠群大小The contig size /bp | 4 000 885 |
| N50支架大小 Scaffold N50 size /bp | 130 035 |
| N50重叠群大小 Contig N50 size /bp | 130 035 |
| N90 支架大小Scaffold N90 size /bp | 41 219 |
| N90 重叠群大小Contig N90 size /bp | 41 219 |
| GC含量 GC content /% | 46.24 |
Table 1 The table of genome simple feature statistics
| 基因组组装 Genome assembly | 大小Size |
|---|---|
| 总支架数 Total number of scaffold /个 | 97 |
| 总重叠群数 Total number of contig /个 | 97 |
| 最大长度 The maximum length /bp | 365 023 |
| 最小长度 The Minimum length /bp | 300 |
| 支架大小 The scaffold size /bp | 4 000 885 |
| 重叠群大小The contig size /bp | 4 000 885 |
| N50支架大小 Scaffold N50 size /bp | 130 035 |
| N50重叠群大小 Contig N50 size /bp | 130 035 |
| N90 支架大小Scaffold N90 size /bp | 41 219 |
| N90 重叠群大小Contig N90 size /bp | 41 219 |
| GC含量 GC content /% | 46.24 |
| 菌株 Strain | 基因组大小 Genome size /bp | GC含量 GC content /% |
|---|---|---|
| Bacillus amyloliquefaciens WF02 | 4.03 | 46.50 |
| Bacillus amyloliquefaciens ZJU1 | 4.06 | 46.40 |
| Bacillus amyloliquefaciens YP6 | 4.01 | 45.90 |
| Bacillus amyloliquefaciens ALB79 | 3.98 | 46.40 |
| Bacillus amyloliquefaciens Y14 | 3.96 | 46.40 |
| Bacillus amyloliquefaciens WS-8 | 3.93 | 46.50 |
| Bacillus amyloliquefaciens MT45 | 3.90 | 46.10 |
| Bacillus amyloliquefaciens KC41 | 4.12 | 46.00 |
| Bacillus amyloliquefaciens T-5 | 4.04 | 46.50 |
| Bacillus amyloliquefaciens LM | 4.00 | 46.24 |
Table 2 Genome comparison between LM and known B. amyloliquefaciens
| 菌株 Strain | 基因组大小 Genome size /bp | GC含量 GC content /% |
|---|---|---|
| Bacillus amyloliquefaciens WF02 | 4.03 | 46.50 |
| Bacillus amyloliquefaciens ZJU1 | 4.06 | 46.40 |
| Bacillus amyloliquefaciens YP6 | 4.01 | 45.90 |
| Bacillus amyloliquefaciens ALB79 | 3.98 | 46.40 |
| Bacillus amyloliquefaciens Y14 | 3.96 | 46.40 |
| Bacillus amyloliquefaciens WS-8 | 3.93 | 46.50 |
| Bacillus amyloliquefaciens MT45 | 3.90 | 46.10 |
| Bacillus amyloliquefaciens KC41 | 4.12 | 46.00 |
| Bacillus amyloliquefaciens T-5 | 4.04 | 46.50 |
| Bacillus amyloliquefaciens LM | 4.00 | 46.24 |
| Cluster | Function category |
|---|---|
| 3 | Bacilysin biosynthetic(细菌溶素生物合成) |
| 4 | Bacillibactin biosynthetic(杆菌素生物合成) |
| 20 | Macrolactin H biosynthetic(大环内酰亚胺H生物合成) |
| 9/23/37 | Difficidin biosynthetic(大环内酯类抗生素生物合成) |
| 1 | Bacillaene biosynthetic(杆菌烯生物合成) |
| 1 | Bacillomycin D biosynthetic(杆菌霉素D生物合成) |
| 2 | Plantazolicin biosynthetic(车前唑啉生物合成) |
| 28 | Plipastatin biosynthetic(巴斯他汀生物合成) |
| 10/33/41 | Surfactin biosynthetic(表面活性素生物合成) |
| 30/38/42 | Fengycin biosynthetic(芬荠素生物合成) |
| 16 | Butirosin A biosynthetic(丁酸普菌素A生物合成) |
Table 3 Analysis of secondary metabolism gene cluster of strain
| Cluster | Function category |
|---|---|
| 3 | Bacilysin biosynthetic(细菌溶素生物合成) |
| 4 | Bacillibactin biosynthetic(杆菌素生物合成) |
| 20 | Macrolactin H biosynthetic(大环内酰亚胺H生物合成) |
| 9/23/37 | Difficidin biosynthetic(大环内酯类抗生素生物合成) |
| 1 | Bacillaene biosynthetic(杆菌烯生物合成) |
| 1 | Bacillomycin D biosynthetic(杆菌霉素D生物合成) |
| 2 | Plantazolicin biosynthetic(车前唑啉生物合成) |
| 28 | Plipastatin biosynthetic(巴斯他汀生物合成) |
| 10/33/41 | Surfactin biosynthetic(表面活性素生物合成) |
| 30/38/42 | Fengycin biosynthetic(芬荠素生物合成) |
| 16 | Butirosin A biosynthetic(丁酸普菌素A生物合成) |
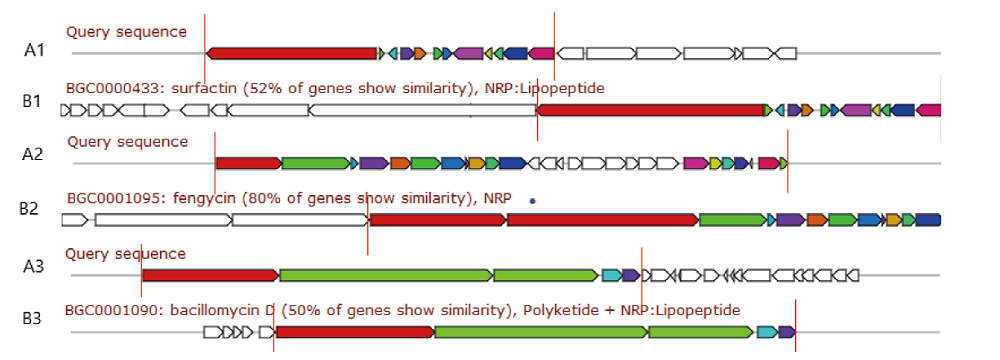
Fig. 5 Comparison of gene cluster structure of three secondary metabolites A1:surfactin gene cluster structure of LM strain;B1:the structure of surfactin gene cluster of FZB42;A2:fengycin gene cluster structure of LM strain;B2:fengycin gene cluster structure of FZB42 strain;A3:bacillomycin gene cluster structure of LM strain;B3:bacillomycin gene cluster structure of FZB42 strain
| 出峰时间 Retention time /min | 分子质量 Molecular mass(m/z) | 离子结形式 Parention | 脂肽类型 Lipopeptide type |
|---|---|---|---|
| 19.768-20.760 | 1042.8(A) | [M+H]+ | C14 iturin A(bacillomycin D)[ |
| 21.669-21.965 | 1056.8(B) | [M+H]+ | C15 iturin A(bacillomycin D)[ |
| 24.291-24.535 | 1070.6(C) | [M+H]+ | C16 iturin A(bacillomycin D)[ |
| 30.213-30.692 | 1480.4(D) | [M+H]+ | C17 fengycin A或C15 fengycin B[ |
| 30.927-32.067 | 1495.0(E) | [M+H]+ | C18 fengycin A或C16 fengycin B[ |
| 32.668-33.550 | 1509.8(F) | [M+H]+ | C17 fengycin B[ |
| 33.663-34.158 | 1524.0(G) | [M+H]+ | C18 fengycin B[ |
| 53.480-54.469 | 1007.8(H) | [M+H]+ | C13 surfactin A或C14 surfactin B或C13 surfactin C[ |
| 57.251-58.102 | 1022.0(I) | [M+H]+ | C14 surfactin A或C15 surfactin B或C14 surfactin C[ |
| 59.318-60.360 | 1036.0(J) | [M+H]+ | C15 surfactin A或C16 surfactin B或C15 surfactin C[ |
Table 4 Analysis results of fermentation broth UPLC-Q-TOF-MS
| 出峰时间 Retention time /min | 分子质量 Molecular mass(m/z) | 离子结形式 Parention | 脂肽类型 Lipopeptide type |
|---|---|---|---|
| 19.768-20.760 | 1042.8(A) | [M+H]+ | C14 iturin A(bacillomycin D)[ |
| 21.669-21.965 | 1056.8(B) | [M+H]+ | C15 iturin A(bacillomycin D)[ |
| 24.291-24.535 | 1070.6(C) | [M+H]+ | C16 iturin A(bacillomycin D)[ |
| 30.213-30.692 | 1480.4(D) | [M+H]+ | C17 fengycin A或C15 fengycin B[ |
| 30.927-32.067 | 1495.0(E) | [M+H]+ | C18 fengycin A或C16 fengycin B[ |
| 32.668-33.550 | 1509.8(F) | [M+H]+ | C17 fengycin B[ |
| 33.663-34.158 | 1524.0(G) | [M+H]+ | C18 fengycin B[ |
| 53.480-54.469 | 1007.8(H) | [M+H]+ | C13 surfactin A或C14 surfactin B或C13 surfactin C[ |
| 57.251-58.102 | 1022.0(I) | [M+H]+ | C14 surfactin A或C15 surfactin B或C14 surfactin C[ |
| 59.318-60.360 | 1036.0(J) | [M+H]+ | C15 surfactin A或C16 surfactin B或C15 surfactin C[ |
| [1] | 梁晓琳, 孙莉, 张娟, 等. 利用 Bacillus amyloliquefaciensSQR9 研制复合微生物肥料[J]. 土壤, 2015, 47(3):558-563. |
| Liang XL, Sun L, Zhang J, et al. Utilization of Bacillus amyloliquefaciensSQR9 for developing compound microbial fertilizer[J]. Soils, 2015, 47(3):558-563. | |
| [2] | 张楠, 吴凯, 沈怡斐, 等. 根际益生菌解淀粉芽孢杆菌SQR9在香蕉根表的定殖行为研究[J]. 南京农业大学学报, 2014, 37(6):59-65. |
| Zhang N, WU K, Shen YF, et al. Investigation of the colonization patterns of plant growth-promoting rhizobacteria Bacillus amyloliquefaciensSQR9 on banana roots[J]. Journal of Nanjing Agricultural University, 2014, 37(6):59-65. | |
| [3] | 杜丽华, 张维瑞, 袁王俊. 解淀粉芽孢杆菌次生代谢产物的研究进展[J]. 科技资讯, 2018, 16(6):128-130. |
| Du LH, Zhang WR, Yuan WJ. Research progress of secondary metabolites of Bacillus amyloliquefaciens[J]. Science and Technology Information, 2018, 16(6):128-130. | |
| [4] | 高学文, 姚仕义, Huong P, 等. 枯草芽孢杆菌B2菌株产生的抑菌活性物质分析[J]. 中国生物防治, 2003, 19(4):175-179. |
| Gao XW, Yao SY, Huong P, et al. Analysis of antibacterial substances produced by Bacillus subtilis B2 strain[J]. Chinese Journal of Biological Control, 2003, 19(4):175-179. | |
| [5] | Priest FG, Goodfellow M, Shute LA, et al. Bacillus amyloliquefaciens. sp. nov. nom. rev[J]. International Journal of Systematic and Evolutionary Microbiology, 1987, 37(1):69-71. |
| [6] | 夏京津, 陈建武, 宋怿, 等. 解淀粉芽孢杆菌HE活性成分鉴定及抗菌特性分析[J]. 南方水产科学, 2019, 15(3):41-49. |
| Xia JJ, Chen JW, Song Y, et al. Identification of antibacterial substances from Bacillus amyloliquefaciens HE and analysis of antibacterial characteristics[J]. Southern Fisheries Science, 2019, 15(3):41-49. | |
| [7] | 戚家明, 孙杉杉, 张东旭 等. 芽孢杆菌BS-6基于全基因组数据的分类鉴定及拮抗能力分析[J]. 生物技术通报, 2019, 35(10):111-118. |
| Qi JM, Sun SS, Zhang DG, et al. Identification and biocontrol activity analysis of Bacillus sp. BS-6 based on genome-wide data[J]. Biotechnology Bulletin, 2019, 35(10):111-118. | |
| [8] |
Arima K, Kakinuma A, Tamura G. Surfactin a crystalline peptidelipid surfactant produced by Bacillus subtilis:isolation characterization and its inhibition of fibrin clot formation[J]. Biochem Biophys Res Commun, 1968, 31(3):488-494.
doi: 10.1016/0006-291X(68)90503-2 URL |
| [9] |
Kakinuma A, Sugino H, Isono M, et al. Determination of fatty acid in surfactin and elucidation of the total structure of surfactin[J]. Agr Biol Chem, 1969, 33(6):973-976.
doi: 10.1080/00021369.1969.10859409 URL |
| [10] | 王志远, 吴兴兴, 吴毅歆, 等. 解淀粉芽孢杆菌B9601-Y2抗性基因标记及其在作物根部的定殖能力[J]. 华中农业大学学报, 2012, 31(3):313-319. |
| Wang ZY, Wu XX, Wu YX, et al. Resistance genes labeling and colonization ability of a biocontrol agent B9601-Y2 of Bacillus amyloliquefaciens in crop rhizospheres[J]. Journal of Huazhong Agricultural University, 2012, 31(3):313-319. | |
| [11] |
Nguyen ATV, Nguyen DV, Tran MT, et al. Isolation and characterization of Bacillus subtilis CH16 strain from chicken gastrointestinal tracts for use as a feed supplement to promote weight gain in broilers[J]. Letters in Applied Microbiology, 2015, 60(6):580-588.
doi: 10.1111/lam.12411 pmid: 25754534 |
| [12] |
Kim BJ, Lee SH, Lyu MA, et al. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene(rpoB)[J]. Journal of Clinical Microbiology, 1999, 37(6):1714.
pmid: 10325313 |
| [13] | Wang LT, Lee FL, Tai CJ, et al. Comparison of gyrB gene sequences, 16S rRNA gene sequences and DNA-DNA hybridization in the Bacillus subtilis group[J]. International Journal of Systematic and Volutionary Microbiology, 2007, 57(8):1846-1850. |
| [14] | 陈成, 崔堂兵, 于平儒. 一株抗真菌的解淀粉芽孢杆菌的鉴定及其抗菌性研究[J]. 现代食品科技, 2011, 27(1):36-39. |
| Chen C, Cui TB, Yu PR. Identification of an anti-fungal strain of Amyloliquefaciens Bacillus and the properties of the antifungal substance[J]. Modern Food Science and Technology, 2011, 27(1):36-39. | |
| [15] | 王继华, 徐世强, 张木清. 解淀粉芽孢杆菌的研究进展[J]. 亚热带农业研究, 2017, 13(3):191-195. |
| Wang JH, Xu SG, Zhang MQ. Research progress on Bacillus amyloliquefaciens[J]. Subtropical Agricultural Research, 2017, 13(3):191-195. | |
| [16] | 李雪飞, 郭丽琼, 叶志伟, 等. 芒果源植物乳杆菌FMNP01的全基因组测序及序列分析[J]. 中国食品学报, 2018, 18(5):232-238. |
| Li XF, Guo LQ, Ye ZW, et al. Whole-genome sequencing and analysis of Lactobacillus plantarum FMNP01 from fresh mangifera indica L[J]. Journal of Chinese Institute of Food Science and Technology, 2018, 18(5):232-238. | |
| [17] | 贾慧慧, 王超男, 魏艳敏, 等. 解淀粉芽胞杆菌BJ-6基因组测序与抗菌代谢产物合成基因分析[J]. 北京农学院学报, 2020, 35(3):64-68. |
| Jia HH, Wang CN, Wei YM, et al. Whole genome sequences of Bacillus amyloliquefaciens BJ-6 and analysis of biosynjournal genes of antibiotic metabolite[J]. Journal of Beijing University of Agriculture, 2020, 35(3):64-68. | |
| [18] |
Koumoutsi A, Chen XH, Henne A, et al. Structural and functional characterization of gene clusters directing nonribosomal synjournal of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens Strain FZB42[J]. Journal of Bacteriology, 2004, 186(4):1084-1096.
doi: 10.1128/JB.186.4.1084-1096.2004 URL |
| [19] | 陈华, 袁成凌, 蔡克周, 等. 枯草芽孢杆菌JA产生的脂肽类抗生素-iturin A的纯化及电喷雾质谱鉴定[J]. 微生物学报, 2008(1):116-120. |
| Chen H, Yuan CL, Cai KZ, et al. Purification and identification of iturin A from Bacillus subtilis JA by electrospray ionization mass spectrometry[J]. Acta Microbiologica Sinica, 2008(1):116-120. | |
| [20] | 董伟欣, 李宝庆, 李社增, 等. 脂肽类抗生素 fengycin 在枯草芽胞杆菌 NCD-2 菌株抑制番茄灰霉病菌中的功能分析[J]. 植物病理学报, 2013, 43(4):401-410. |
| Dong WX, Li BQ, Li SZ, et al. The fengycin lipopeptides are major components in the Bacillus subtilis strain NCD-2 against the growth of Botrytis cinereal[J]. Acta Phytopathologica Sinica, 2013, 43(4):401-410. | |
| [21] | 张荣胜, 王晓宇, 罗楚平, 等. 解淀粉芽孢杆菌Lx-11产脂肽类物质鉴定及表面活性素对水稻细菌性条斑病的防治作用[J]. 中国农业科学, 2013, 46(10):2014-2021. |
| Zhang RS, Wang XY, Luo CP, et al. Identification of the lipopeptides from Bacillus amyloliquefaciensLx-11 and biocontrol efficacy of surfactin against bacterial leaf streak[J]. Scientia Agricultura Sinica, 2013, 46(10):2014-2021. | |
| [22] | 胡陈云, 李勇, 刘敏, 等. 枯草芽孢杆菌ge25对两种人参病原菌的抑制作用及脂肽类抑菌代谢产物的鉴定[J]. 中国生物防治学报, 2015, 31(3):386-393. |
| Hu CY, Li Y, Liu M, et al. Antagonism of Bacillus subtilis ge25 against two kinds of ginseng pathogens and identification of antifungal lipopeptide metabolites[J]. Chinese Journal of Biological Control, 2015, 31(3):386-393. | |
| [23] | 胡陈云, 李勇, 刘敏, 等. 枯草芽孢杆菌ge25菌株发酵及其抑菌活性成分的初步研究[J]. 中国中药志, 2014, 39(14):2624-2628. |
| Hu CY, Li Y, Liu M, et al. Fermentation of Bacillus subtilis ge25 strain and its antibacterial active components[J]. China Journal of Chinese Materia Medica, 2014, 39(14):2624-2628. |
| [1] | WANG Teng-hui, GE Wen-dong, LUO Ya-fang, FAN Zhen-yu, WANG Yu-shu. Gene Mapping of Kale White Leaves Based on Whole Genome Re-sequencing of Extreme Mixed Pool(BSA) [J]. Biotechnology Bulletin, 2023, 39(9): 176-182. |
| [2] | LI Xue-qi, ZHANG Su-jie, YU Man, HUANG Jin-guang, ZHOU Huan-bin. Establishment of CRISPR/CasX-based Genome Editing Technology in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 40-48. |
| [3] | FANG Lan, LI Yan-yan, JIANG Jian-wei, CHENG Sheng, SUN Zheng-xiang, ZHOU Yi. Isolation, Identification and Growth-promoting Characteristics of Endohyphal Bacterium 7-2H from Endophytic Fungi of Spiranthes sinensis [J]. Biotechnology Bulletin, 2023, 39(8): 272-282. |
| [4] | RAO Zi-huan, XIE Zhi-xiong. Isolation and Identification of a Cellulose-degrading Strain of Olivibacter jilunii and Analysis of Its Degradability [J]. Biotechnology Bulletin, 2023, 39(8): 283-290. |
| [5] | GUO Shao-hua, MAO Hui-li, LIU Zheng-quan, FU Mei-yuan, ZHAO Ping-yuan, MA Wen-bo, LI Xu-dong, GUAN Jian-yi. Whole Genome Sequencing and Comparative Genome Analysis of a Fish-derived Pathogenic Aeromonas Hydrophila Strain XDMG [J]. Biotechnology Bulletin, 2023, 39(8): 291-306. |
| [6] | ZHANG Dao-lei, GAN Yu-jun, LE Liang, PU Li. Epigenetic Regulation of Yield-related Traits in Maize and Epibreeding [J]. Biotechnology Bulletin, 2023, 39(8): 31-42. |
| [7] | SHI Jia-xin, LIU Kai, ZHU Jin-jie, QI Xian-tao, XIE Chuan-xiao, LIU Chang-lin. Gene Editing Reshaping Maize Plant Type for Increasing Hybrid Yield [J]. Biotechnology Bulletin, 2023, 39(8): 62-69. |
| [8] | DU Dong-dong, QIAN Jing, LI Si-qi, LIU Wen-fei, WEI Xiang-li, LIU Chang-yong, LUO Rui-feng, KANG Li-chao. Whole Genome Sequencing and Analysis of Listeria monocytogenes Strain LMXJ15 [J]. Biotechnology Bulletin, 2023, 39(7): 298-306. |
| [9] | LI Yu-zhen, MEI Tian-xiu, LI Zhi-wen, WANG Qi, LI Jun, ZOU Yue, ZHAO Xin-qing. Advances in Genomic Studies and Metabolic Engineering of Red Yeasts [J]. Biotechnology Bulletin, 2023, 39(7): 67-79. |
| [10] | YOU Zi-juan, CHEN Han-lin, DENG Fu-cai. Research Progress in the Extraction and Functional Activities of Bioactive Peptides from Fish Skin [J]. Biotechnology Bulletin, 2023, 39(7): 91-104. |
| [11] | YIN Ming-hua, YU Huan-yuan, XIAO Xin-yi, WANG Yu-ting. Chloroplast Genomic Characterization and Phylogenetic Analysis of Colocasia esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan [J]. Biotechnology Bulletin, 2023, 39(6): 233-247. |
| [12] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [13] | LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit [J]. Biotechnology Bulletin, 2023, 39(4): 136-147. |
| [14] | ZHOU Xiao-jie, YANG Si-qi, ZHANG Yi-wen, XU Jia-qi, YANG Sheng. CRISPR-associated Transposases and Their Applications in Bacterial Genome Editing [J]. Biotechnology Bulletin, 2023, 39(4): 49-58. |
| [15] | XIAO Xiao-jun, CHEN Ming, HAN De-peng, YU Pao-lan, ZHENG Wei, XIAO Guo-bin, ZHOU Qing-hong, ZHOU Hui-wen. Genome Wide Association Analysis of Thousand Seed Weight in Brassica napus L. [J]. Biotechnology Bulletin, 2023, 39(3): 143-151. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||