








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (10): 100-109.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0106
Previous Articles Next Articles
TIAN Geng( ), GAO Wei-qiang, CHEN Xiao-bo, ZHANG Chun-xiao(
), GAO Wei-qiang, CHEN Xiao-bo, ZHANG Chun-xiao( )
)
Received:2021-01-27
Online:2021-10-26
Published:2021-11-12
Contact:
ZHANG Chun-xiao
E-mail:741033155@qq.com;cxzhang2009@163.com
TIAN Geng, GAO Wei-qiang, CHEN Xiao-bo, ZHANG Chun-xiao. Directed Mutagenesis of β-mannanase Gene from Bacillus licheniformis KD-1 for Improving Enzyme Activity and Stability[J]. Biotechnology Bulletin, 2021, 37(10): 100-109.
| 质粒Plasmids | 备注Notes | 来源Source |
|---|---|---|
| pWB980 | [20] | |
| pWB-manBl | pWB980 with manBl gene | 本研究This study |
| pWB-manBl(K291E) | pWB980 with manBl gene mutant K291E | 本研究This study |
| pWB-manBl(T112R) | pWB980 with manBl gene mutant T112R | 本研究This study |
| pWB-manBl(L211I) | pWB980 with manBl gene mutant L211I | 本研究This study |
| pWB-manBl- T112R/K291E | pWB980 with manBl gene mutant T112R/K291E | 本研究This study |
| pWB-manBl- L211I/ K291E | pWB980 with manBl gene mutant L211I/K291E | 本研究This study |
| pWB-manBl- T112R/L211I | pWB980 with manBl gene mutant T112R/L211I | 本研究This study |
| pWB-manBl-His | pWB980 with manBl gene fused with 6*His tagged at C terminal | 本研究This study |
| pWB-manBl(T112R/K291E)-His | pWB980 with manBl(T112R/K291E)gene fused with 6*His tagged at C terminal | 本研究This study |
Table 1 Plasmids used in this study
| 质粒Plasmids | 备注Notes | 来源Source |
|---|---|---|
| pWB980 | [20] | |
| pWB-manBl | pWB980 with manBl gene | 本研究This study |
| pWB-manBl(K291E) | pWB980 with manBl gene mutant K291E | 本研究This study |
| pWB-manBl(T112R) | pWB980 with manBl gene mutant T112R | 本研究This study |
| pWB-manBl(L211I) | pWB980 with manBl gene mutant L211I | 本研究This study |
| pWB-manBl- T112R/K291E | pWB980 with manBl gene mutant T112R/K291E | 本研究This study |
| pWB-manBl- L211I/ K291E | pWB980 with manBl gene mutant L211I/K291E | 本研究This study |
| pWB-manBl- T112R/L211I | pWB980 with manBl gene mutant T112R/L211I | 本研究This study |
| pWB-manBl-His | pWB980 with manBl gene fused with 6*His tagged at C terminal | 本研究This study |
| pWB-manBl(T112R/K291E)-His | pWB980 with manBl(T112R/K291E)gene fused with 6*His tagged at C terminal | 本研究This study |
| 菌株 Strain | 备注Notes | 来源Source |
|---|---|---|
| B. subtilis DB104 | his,nprR2,nprE18,ΔaprA3 | [21] |
| B. licheniformis KD-1 | 本研究This study | |
| WB980 | B. subtilis DB104(pWB980),Kanr | 本研究This study |
| MTV13 | B. subtilis DB104(pWB-manBl),Kanr | 本研究This study |
| ABV3 | B. subtilis DB104(pWB-manBl-K291E),Kanr | 本研究This study |
| T2 | B. subtilis DB104(pWB-manBl-T112R),Kanr | 本研究This study |
| L4 | B. subtilis DB104(pWB-manBl-L211I),Kanr | 本研究This study |
| LK2 | B. subtilis DB104(pWB-manBl-L211I /K291E),Kanr | 本研究This study |
| TL1 | B. subtilis DB104(pWB-manBl-T112R/L211I),Kanr | 本研究This study |
| TK7 | B. subtilis DB104(pWB-manBl-T112R/K291E),Kanr | 本研究This study |
| A3 | B. subtilis DB104(pWB-manBl-His6),Kanr | 本研究This study |
| C5 | B. subtilis DB104(pWB-manBl-T112R/L211I-His6),Kanr | 本研究This study |
Table 2 Strains used in this study
| 菌株 Strain | 备注Notes | 来源Source |
|---|---|---|
| B. subtilis DB104 | his,nprR2,nprE18,ΔaprA3 | [21] |
| B. licheniformis KD-1 | 本研究This study | |
| WB980 | B. subtilis DB104(pWB980),Kanr | 本研究This study |
| MTV13 | B. subtilis DB104(pWB-manBl),Kanr | 本研究This study |
| ABV3 | B. subtilis DB104(pWB-manBl-K291E),Kanr | 本研究This study |
| T2 | B. subtilis DB104(pWB-manBl-T112R),Kanr | 本研究This study |
| L4 | B. subtilis DB104(pWB-manBl-L211I),Kanr | 本研究This study |
| LK2 | B. subtilis DB104(pWB-manBl-L211I /K291E),Kanr | 本研究This study |
| TL1 | B. subtilis DB104(pWB-manBl-T112R/L211I),Kanr | 本研究This study |
| TK7 | B. subtilis DB104(pWB-manBl-T112R/K291E),Kanr | 本研究This study |
| A3 | B. subtilis DB104(pWB-manBl-His6),Kanr | 本研究This study |
| C5 | B. subtilis DB104(pWB-manBl-T112R/L211I-His6),Kanr | 本研究This study |
| 引物名称 Primer name | 引物序列Prime sequence (5'-3') |
|---|---|
| Bl man2-F1 | gtgaaaaaaaRcatcgtttg |
| BL man2-R1 | ttattccacRacaggcgtcaaag |
| BL man2-F2 | CACACCGTTTCTCCGGTGAAC |
| manBT-2 | GTAAGTCCCGTCTAGCCTTGCCCTTATTCCACAACAGGCGTCAAAG |
| amyT-F | TGTTGTGGAATAAGGGCAAGGCTAGACGGGACTTAC |
| amyT-R | CTAAATCGTGTCTTTCTTGGAACTTCCAGGGTATGTTTCTCTTTGATGTC |
| spmanB-F | AGGCGCAACTCAAGCTTTTGCCCACACCGTTTCTCCGGTGAAC |
| Pvf-1 | ACATCAAAGAGAAACATACCCTGGAAGTTCCAAGAAAGACACGATTTAG |
| Pvf-2 | GTTCACCGGAGAAACGGTGTGGGCAAAAGCTTGAGTTGCGCCT |
| R-manBl-F | GTTTCTCCGGTGAACCCGAATGCCCA |
| R-manBl-R | CGCCGTCCCATATCTCTCCTTTATTCA |
| V-manBl-F | TGAATAAAGGAGAGATATGGGACGGCG |
| V-manBl-R | TGGGCATTCGGGTTCACCGGAGAAAC |
| DR-manBl-R | GAAGTACGTCGTTTCCGGGTATTTTTCTTTG |
| DR-manBl-F | CAAAGAAAAATACCCGGAAACGACGTACTTC |
| BL-L211I-R | CGCATACACCCACAAGATATGATCCAGGCCTCT |
| BL-L211I-F | AGAGGCCTGGATCATATCTTGTGGGTGTATGCG |
| BL-T112R-R | ATAATGACCCGATCTAAACGCGGGGTTTGCGAG |
| BL-T112R-F | CTCGCAAACCCCGCGTTTAGATCGGGTCATTAT |
| V-His-F | CACCATCACCATCACCATTAAGGGCAAGGCTAGACGGGACTTAC |
| V-manBl-R | TGGGCATTCGGGTTCACCGGAGAAAC |
| R-manBl-F | GTTTCTCCGGTGAACCCGAATGCCCA |
| manBl-His-R | TTGCCCTTAATGGTGATGGTGATGGTGTTCCACAACAGGCGTCAAAG |
Table 3 Primes used in this study
| 引物名称 Primer name | 引物序列Prime sequence (5'-3') |
|---|---|
| Bl man2-F1 | gtgaaaaaaaRcatcgtttg |
| BL man2-R1 | ttattccacRacaggcgtcaaag |
| BL man2-F2 | CACACCGTTTCTCCGGTGAAC |
| manBT-2 | GTAAGTCCCGTCTAGCCTTGCCCTTATTCCACAACAGGCGTCAAAG |
| amyT-F | TGTTGTGGAATAAGGGCAAGGCTAGACGGGACTTAC |
| amyT-R | CTAAATCGTGTCTTTCTTGGAACTTCCAGGGTATGTTTCTCTTTGATGTC |
| spmanB-F | AGGCGCAACTCAAGCTTTTGCCCACACCGTTTCTCCGGTGAAC |
| Pvf-1 | ACATCAAAGAGAAACATACCCTGGAAGTTCCAAGAAAGACACGATTTAG |
| Pvf-2 | GTTCACCGGAGAAACGGTGTGGGCAAAAGCTTGAGTTGCGCCT |
| R-manBl-F | GTTTCTCCGGTGAACCCGAATGCCCA |
| R-manBl-R | CGCCGTCCCATATCTCTCCTTTATTCA |
| V-manBl-F | TGAATAAAGGAGAGATATGGGACGGCG |
| V-manBl-R | TGGGCATTCGGGTTCACCGGAGAAAC |
| DR-manBl-R | GAAGTACGTCGTTTCCGGGTATTTTTCTTTG |
| DR-manBl-F | CAAAGAAAAATACCCGGAAACGACGTACTTC |
| BL-L211I-R | CGCATACACCCACAAGATATGATCCAGGCCTCT |
| BL-L211I-F | AGAGGCCTGGATCATATCTTGTGGGTGTATGCG |
| BL-T112R-R | ATAATGACCCGATCTAAACGCGGGGTTTGCGAG |
| BL-T112R-F | CTCGCAAACCCCGCGTTTAGATCGGGTCATTAT |
| V-His-F | CACCATCACCATCACCATTAAGGGCAAGGCTAGACGGGACTTAC |
| V-manBl-R | TGGGCATTCGGGTTCACCGGAGAAAC |
| R-manBl-F | GTTTCTCCGGTGAACCCGAATGCCCA |
| manBl-His-R | TTGCCCTTAATGGTGATGGTGATGGTGTTCCACAACAGGCGTCAAAG |
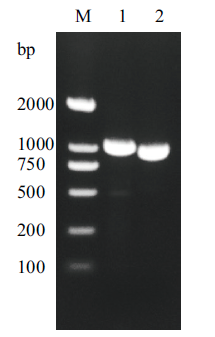
Fig. 1 The β-mannanase gene PCR product M:DNA marker, Lane 1:the complete β-mannanase gene; Lane 2:the β-mannanase gene without signal peptide sequence
| Enzyme | Km/(mg·mL-1) | Vmax/(µg·s-1) |
|---|---|---|
| ManBl | 3.80 | 0.499 |
| ManBl(K291E) | 4.80 | 0.879 |
| ManBl(L211I) | 5.22 | 0.382 |
| ManBl(T112R) | 4.11 | 0.711 |
| ManBl(T112R/L211I) | 5.18 | 0.951 |
| ManBl(L211I/K291E) | 6.46 | 0.793 |
| ManBl(T112R/K291E) | 2.67 | 0.635 |
Table 4 Kinetic parateters of ManBl and its variants
| Enzyme | Km/(mg·mL-1) | Vmax/(µg·s-1) |
|---|---|---|
| ManBl | 3.80 | 0.499 |
| ManBl(K291E) | 4.80 | 0.879 |
| ManBl(L211I) | 5.22 | 0.382 |
| ManBl(T112R) | 4.11 | 0.711 |
| ManBl(T112R/L211I) | 5.18 | 0.951 |
| ManBl(L211I/K291E) | 6.46 | 0.793 |
| ManBl(T112R/K291E) | 2.67 | 0.635 |
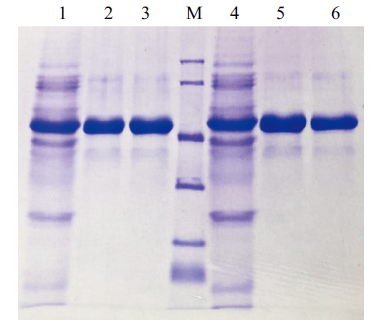
Fig. 5 Protein purification results Lane 1: the fermentation broth of B. subtilis(pWB-manBl-His). Lane 2 and 3: The purified manBl-His. Lane 4: The fermentation broth of B. subtilis(pWB-manBl(T112R/K291E)-His); Lane 5 and 6: The purified manBl(T112R/K291E)-His
| [1] |
Songsiriritthigul C, Buranabanyat B, Haltrich D, et al. Efficient recombinant expression and secretion of a thermostable GH26 mannan endo-1, 4-β-mannosidase from Bacillus licheniformis in Escherichia coli[J]. Microb Cell Factories, 2010, 9(4):20-32.
doi: 10.1186/1475-2859-9-20 URL |
| [2] |
Zhang W, Liu Z, Zhou S, et al. Cloning and expression of a β-mannanase gene from Bacillus sp. MK-2 and its directed evolution by random mutagenesis[J]. Enzyme Microb Technol, 2019, 124(5):70-78.
doi: 10.1016/j.enzmictec.2019.02.003 URL |
| [3] |
Dawood A, Ma K. Applications of microbial β-mannanases[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8:1-17.
doi: 10.3389/fbioe.2020.00001 URL |
| [4] |
Wu G, Bryant MM, Voitle RA, et al. Effects of β-mannanase in corn-soy diets on commercial leghorns in second-cycle hens[J]. Poult Sci, 2005, 84(6):894-897.
doi: 10.1093/ps/84.6.894 URL |
| [5] | 杨少杰, 高海有, 李晞, 等. 甘露聚糖酶和木聚糖酶在纸浆漂白中的应用[J]. 造纸科学与技术, 2016, 35(4):71-76. |
| Yang SJ, Gao HY, Li X, et al. Application of mannanase and xylanase in the pulp bleaching[J]. Paper Science&Technology, 2016, 35(4):71-76. | |
| [6] |
Singh S, Singh G, Khatri M, et al. Thermo and alkali stable β-mannanase:Characterization and application for removal of food(mannans based)stain[J]. International J Biol Macromol, 2019, 134:536-546.
doi: 10.1016/j.ijbiomac.2019.05.067 URL |
| [7] | 崔婷婷, 刘锐, 吴涛, 等. 酶解魔芋葡甘聚糖对冷冻面团拉伸特性的影响[J]. 食品工业科技, 2019, 40(20):7-15. |
| Cui TT, Liu R, Wu T, et al. Effects of Enzymatic hydrolysis of konjac glucomannan on tensile properties of frozen dough[J]. Science and Technology of Food Industry, 2019, 40(20):7-15. | |
| [8] |
Chauhan PS, Sharma P, Puri N, et al. Purification and characterization of an alkali-thermostable β-mannanase from Bacillus nealsonii PN-11 and its application in mannooligosaccharides preparation having prebiotic potential[J]. Eur Food Res Technol, 2014, 238(2):927-936.
doi: 10.1007/s00217-014-2170-7 URL |
| [9] |
Sachslehner A, Foidl G, Foidl N, et al. Hydrolysis of isolated coffee mannan and coffee extract by mannanases of Sclerotium rolfsii[J]. J Biotechnol, 2000, 80(2):127-134.
pmid: 10908793 |
| [10] |
Nadaroglu H, Adiguzel A, Adiguzel G. Purification and characterisation of β-mannanase from Lactobacillus plantarum(M24)and its applications in some fruit juices[J]. Int J Food Sci Tech, 2015, 50(5):1158-1165.
doi: 10.1111/ijfs.2015.50.issue-5 URL |
| [11] |
Adiguzel A, Nadaroglu H, Adiguzel G. Purification and characterization of β-mannanase from Bacillus pumilus(M27)and its applications in some fruit juices[J]. J Food Sci Tech, 2014, 52(8):5292-5298.
doi: 10.1007/s13197-014-1609-y URL |
| [12] |
Srivastava PK, Kapoor M. Production, properties, and applications of endo-β-mannanases[J]. Biotechnol Adv, 2017, 35(1):1-19.
doi: S0734-9750(16)30136-7 pmid: 27836790 |
| [13] |
Rahmani N, Kashiwagi N, Lee JM, et al. Mannan endo-1, 4-β-mannosidase from Kitasatospora sp. isolated in Indonesia and its potential for production of mannooligosaccharides from mannan polymers[J]. AMB Expr, 2017, 7:100-110.
doi: 10.1186/s13568-017-0401-6 URL |
| [14] |
Mano MCR, Neri-Numa IA, Silva JB, et al. Oligosaccharide biotechnology:an approach of prebiotic revolution on the industry[J]. Appl Microbiol Biotechnol, 2018, 102(1):17-37.
doi: 10.1007/s00253-017-8564-2 URL |
| [15] |
Wang H, Zhang X, Wang X, et al. Mannan-oligosaccharide modulates the obesity and gut microbiota in high-fat diet-fed mice[J]. Food Funct, 2018, 9(7):3916-3929.
doi: 10.1039/C8FO00209F URL |
| [16] |
Katrolia P, Zhou P, Zhang P, et al. High level expression of a novel β-mannanase from Chaetomium sp. exhibiting efficient mannan hydrolysis[J]. Carbohydr Polym, 2012, 87:480-490.
doi: 10.1016/j.carbpol.2011.08.008 URL |
| [17] |
Song Y, Fu G, Dong H, et al. High-Efficiency secretion of β-mannanase in Bacillus subtilis through protein synjournal and secretion optimization[J]. J Agric Food Chem, 2017, 65(12):2540-2548.
doi: 10.1021/acs.jafc.6b05528 URL |
| [18] | Songsiriritthigul C, Lapboonrueng S, Roytrakul S, et al. Crystallization and preliminary crystallographic analysis of β-mannanase from Bacillus licheniformis[J]. Acta Cryst, 2011, 67(Pt 2):217-220. |
| [19] |
Yan XX, An XM, Gui LL, et al. From structure to function:Insights into the catalytic substrate specificity and thermostability displayed by Bacillus subtilis mannanase bcman[J]. J Mol Biol, 2008, 379(3):535-544.
doi: 10.1016/j.jmb.2008.03.068 URL |
| [20] |
Wu SC, Wong SL. Development of improved pUB110-based vectors for expression and secretion studies in Bacillus subtilis[J]. J Biotechnol, 1999, 72(3):185-195.
doi: 10.1016/S0168-1656(99)00101-7 URL |
| [21] |
Kawamura F, Doi RH. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases[J]. J Bacteriol, 1984, 160(1):442-444.
doi: 10.1128/jb.160.1.442-444.1984 pmid: 6434524 |
| [22] |
Vojcic L, Despotovic D, Martinez R, et al. An efficient transformation method for Bacillus subtilis DB104[J]. Appl Microbiol Biotechnol, 2012, 94:487-493.
doi: 10.1007/s00253-012-3987-2 URL |
| [23] |
You C, Zhang XZ, Zhang YH. Simple cloning via direct transform-ation of PCR product(DNA Multimer)to Escherichia coli and Bacillus subtilis[J]. Appl Environ Microbiol, 2012, 78(5):1593-1595.
doi: 10.1128/AEM.07105-11 URL |
| [24] |
Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar[J]. Analytical Chemistry, 1959, 31(3):426-428.
doi: 10.1021/ac60147a030 URL |
| [25] | 田庚, 高伟强, 陈晓波, 等. 热稳定性高β-甘露聚糖酶产生菌的筛选、鉴定及酶学性质研究[J]. 食品工业科技, 2020, 41(19):127-131. |
| Tian G, Gao WQ, Chen XB, et al. Isolation, identification and enzyme properties of a β-mannanase producing strain with high thermal stability[J]. Science and Technology of Food Industry, 2020, 41(19):127-131. | |
| [26] |
Nielsen H, Tsirigos KD, Brunak S, et al. A brief history of protein sorting prediction[J]. Protein J, 2019, 38(3):200-216.
doi: 10.1007/s10930-019-09838-3 pmid: 31119599 |
| [27] |
Capriotti E, Fariselli P, Casadio R. I-Mutant2. 0:predicting stability changes upon mutation from the protein sequence or structure[J]. Nucleic Acids Res, 2005, 33(suppl 2):W306-W310.
doi: 10.1093/nar/gki375 URL |
| [28] |
Blum M, Chang H, Chuguransky S, et al. The InterPro protein families and domains database:20 years on[J]. Nucleic Acids Res, 2021, 49(D1):D344-D354.
doi: 10.1093/nar/gkaa977 URL |
| [29] |
Wang XC, You SP, Zhang JX, et al. Rational design of a thermophilic β-mannanase from Bacillus subtilis TJ-102 to improve its thermostability[J]. Enzyme and Microbial Technology, 2018, 118:50-56.
doi: 10.1016/j.enzmictec.2018.07.005 URL |
| [30] | Kweun MA, Lee MS, Choi JH, et al. Cloning of a Bacillus subtilis WL-7 mannanase gene and characterization of the gene product[J]. J Microbiol Biotechnol, 2004, 14(6):1295-1302. |
| [31] |
Zhou C, Xue Y, Ma Y. Characterization and high-efficiency secreted expression in Bacillus subtilis of a thermo-alkaline β-mannanase from an alkaliphilic Bacillus clausii strain S10[J]. Microb Cell Fact, 2018, 17(1):124-142.
doi: 10.1186/s12934-018-0973-0 URL |
| [1] | YAN Tao, CHEN Ke-ke, YANG Heng-fei, ZHU Jian-guo, XIA Jiu-xue, FANG Shu-guang. Study on Factors Affecting the Storage Survival Rates of Probiotic Bacteria Powder [J]. Biotechnology Bulletin, 2023, 39(4): 296-303. |
| [2] | SONG Hai-na, WU Xin-tong, YANG Lu-yu, GENG Xi-ning, ZHANG Hua-min, SONG Xiao-long. Selection and Validation of Reference Genes for RT-qPCR in Allium tuberosum Infected by Botrytis squamosa [J]. Biotechnology Bulletin, 2023, 39(3): 101-115. |
| [3] | ZHU Ying-xuan, LI Ke-jing, HE Min, ZHENG Dao-qiong. Research Progress in the Exploring Genomic Variations Driven by Stress Factors Using the Yeast Model [J]. Biotechnology Bulletin, 2023, 39(11): 191-204. |
| [4] | YANG Jun-zhao, ZHANG Xin-rui, SUN Qing-yang, ZHENG Fei. Affecting Mechanism of Loop B3 on the Function of GH7 Endoglucanase [J]. Biotechnology Bulletin, 2023, 39(10): 281-291. |
| [5] | LI Sheng-yan, LI Xiang-yin, LI Peng-cheng, ZHANG Ming-jun, ZHANG Jie, LANG Zhi-hong. Identification of Target Traits and Genetic Stability of Transgenic Maize 2HVB5 [J]. Biotechnology Bulletin, 2023, 39(1): 21-30. |
| [6] | ZHAO Zhong-juan, YANG Kai, HU Jin-dong, WEI Yan-li, LI Ling, XU Wei-sheng, LI Ji-shun. Effects of Trichoderma harzianum ST02 on the Growth of Peppermint and Physicochemical Properties of Root Zone Soil Under Salt Stress [J]. Biotechnology Bulletin, 2022, 38(7): 224-235. |
| [7] | YUAN Cun-xia, LI Yan-nan, ZHANG Xiao-chong, YANG Rui, LIU Jian-li, LI Jing-yu. Physiological and Biochemical Response Characteristics of Bacillus sp. ZJS3 Under As3+ Stress [J]. Biotechnology Bulletin, 2022, 38(7): 236-246. |
| [8] | ZHU Qiu-yu, DUAN Xu-guo. Recombinant Expression and Site-directed Mutagenesis of L-aspartate-α-decarboxylase,and the Establishment of High-throughput Assay Method [J]. Biotechnology Bulletin, 2022, 38(5): 269-278. |
| [9] | WANG Zi-yan, WANG Jian, ZHANG Lun, GUI Shui-qing, LU Xue-mei. Study on Antibacterial Stability of Musca domestica Cecropin-MDC Against Salmonella typhimurium [J]. Biotechnology Bulletin, 2022, 38(3): 149-156. |
| [10] | WANG Xiao-qin, HUANG Yin-ping, WANG Wei-qian, WU Ping, QUAN Shu. Expression and Purification of the MLL3SET Protein with a Site-directed Mutation of an Unnatural Amino Acid [J]. Biotechnology Bulletin, 2022, 38(3): 194-202. |
| [11] | LIANG Xing-xing, WANG Jia, XU Wen-tao. Research Progress in Phosphorylation Modification of Antiviral Nucleotide Analogs [J]. Biotechnology Bulletin, 2022, 38(2): 218-226. |
| [12] | JIA Hai-hong, LI Bing-qing. Research Progress in the Post-translational Modification of Superoxide Dismutase [J]. Biotechnology Bulletin, 2022, 38(2): 237-244. |
| [13] | ZHANG Chen, ZHANG Tong-tong, LIU Hai-ping. Screening and Identification of Ethylene-forming Enzymes with High Activity and Thermostability [J]. Biotechnology Bulletin, 2022, 38(11): 269-276. |
| [14] | WU Qi-man, TIAN Shi-han, LI Yun-ye, PAN Ying-jie, ZHANG Ying. Effects of Microbial Fertilizer on Cucumis sativus L. Growth,Yield and Quality [J]. Biotechnology Bulletin, 2022, 38(1): 125-131. |
| [15] | JIANG Di, XU Chun-cheng. Research Progress in the Succession of Microbial Communities in Total Mixed Ration Silage [J]. Biotechnology Bulletin, 2021, 37(9): 31-38. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||