








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (11): 97-103.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0114
Previous Articles Next Articles
WANG Shu-xuan1( ), XIANG Gang1, MA Xiao-jing2(
), XIANG Gang1, MA Xiao-jing2( ), YU Jing1(
), YU Jing1( )
)
Received:2022-01-25
Online:2022-11-26
Published:2022-12-01
Contact:
MA Xiao-jing,YU Jing
E-mail:wsh-email@sjtu.edu.cn;xim2002@med.cornell.edu;jingyu@sjtu.edu.cn
WANG Shu-xuan, XIANG Gang, MA Xiao-jing, YU Jing. Construction of Galectin-1 Overexpressing 4T1 Mammary Tumor Cells and Its Effects on the Proliferation and Migration[J]. Biotechnology Bulletin, 2022, 38(11): 97-103.
| 基因 Gene | 引物序列 Primer sequence(5'-3') |
|---|---|
| Lgals1 | F:AACCTGGGGAATGTCTCAAAGT |
| R:GGTGATGCACACCTCTGTGA | |
| Gapdh | F:AGGTCGGTGTGAACGGATTTG |
| R:TGTAGACCATGTAGTTGAGGTCA |
Table 1 Lgals1 and Gapdh primer sequences
| 基因 Gene | 引物序列 Primer sequence(5'-3') |
|---|---|
| Lgals1 | F:AACCTGGGGAATGTCTCAAAGT |
| R:GGTGATGCACACCTCTGTGA | |
| Gapdh | F:AGGTCGGTGTGAACGGATTTG |
| R:TGTAGACCATGTAGTTGAGGTCA |
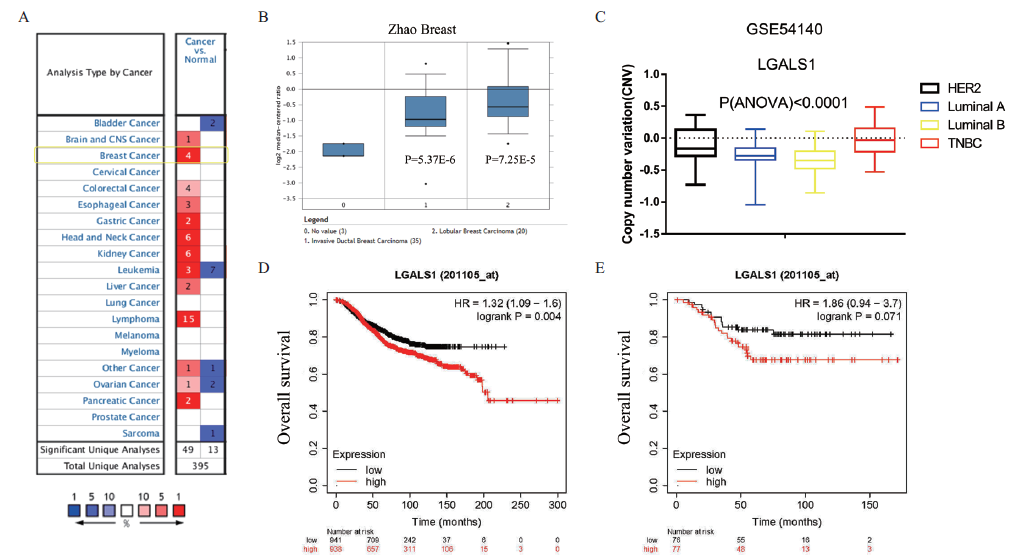
Fig. 1 Bioinformatics database analysis of the expression of galectin-1 in breast cancer and the correlation of it with overall survival rate of breast cancer patients A:Oncomine database analysis of galectin-1 expression in various cancers(The long yellow box indicates breast cancer;the number indicates the amounts of samples that galectin-1 was analyzed in different cancers. Darker red indicates higher gene expression,darker blue indicates lower expression). B:Oncomine database analysis of differential mRNA expression of galectin-1 in normal and tumor tissues in Zhao Breast dataset. C:GEO database(GSE54140 data set)analysis of copy number variation of galectin-1 gene in different breast cancer subtypes. D-E:Kaplan-Meier analysis of the correlation between galectin-1 expression and the overall survival rate of(D)breast cancer patients and(E)TNBC patients
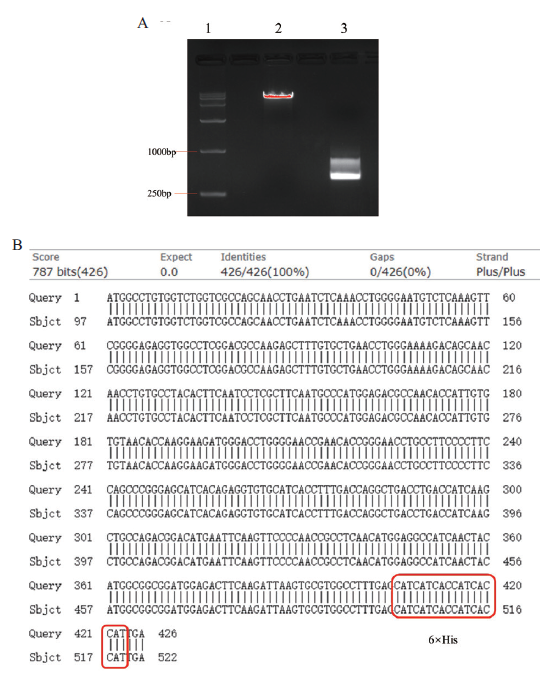
Fig. 2 Digestion and sequencing analysis of the galactin-1-expressed plasmid pCDH-Lgals1 A:Agarose gel electrophoresis of the digestion of pCDH vector and PCR amplification products of Lgals1. 1:15000 DNA ladder;2:pCDH digestion vector;3:Lgals1 PCR products. B:Alignment of DNA sequencing results of galectin-1 in the constructed expression plasmid(the sequence of 6×His tag is shown in red box)

Fig. 3 qPCR and Western Blot analysis of galectin-1 in galectin-1 overexpressing 4T1 cells A:qPCR analysis of mRNA levels of galectin-1 in galectin-1 overexpressing 4T1 cell line. B:Western Blot analysis of protein levels of galectin-1 in galectin-1 overexpressing 4T1 cells. C:Quantitative diagram of the B chart

Fig. 5 Overexpression of galectin-1 promotes the migration and invasion of triple-negative breast cancer 4T1 cells A:Transwell migration assay. B:Transwell invasion assay
| [1] |
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020[J]. CA Cancer J Clin, 2020, 70(1):7-30.
doi: 10.3322/caac.21590 URL |
| [2] |
Liao LQ, Song M, Li X, et al. E3 ubiquitin ligase UBR5 drives the growth and metastasis of triple-negative breast cancer[J]. Cancer Res, 2017, 77(8):2090-2101.
doi: 10.1158/0008-5472.CAN-16-2409 pmid: 28330927 |
| [3] |
Shi JH, Liu FQ, Song YQ. Progress: targeted therapy, immunotherapy, and new chemotherapy strategies in advanced triple-negative breast cancer[J]. Cancer Manag Res, 2020, 12: 9375-9387.
doi: 10.2147/CMAR.S272685 pmid: 33061626 |
| [4] |
Lyons TG. Targeted therapies for triple-negative breast cancer[J]. Curr Treat Options Oncol, 2019, 20(11):82.
doi: 10.1007/s11864-019-0682-x URL |
| [5] | Núñez Abad M, Calabuig-Fariñas S, Lobo de Mena M, et al. Update on systemic treatment in early triple negative breast cancer[J]. Ther Adv Med Oncol, 2021, 13: 1758835920986749. |
| [6] |
Yin L, Duan JJ, Bian XW, et al. Triple-negative breast cancer molecular subtyping and treatment progress[J]. Breast Cancer Res, 2020, 22(1):61.
doi: 10.1186/s13058-020-01296-5 pmid: 32517735 |
| [7] |
Sporikova Z, Koudelakova V, Trojanec R, et al. Genetic markers in triple-negative breast cancer[J]. Clin Breast Cancer, 2018, 18(5):e841-e850.
doi: 10.1016/j.clbc.2018.07.023 URL |
| [8] |
Hirbe AC, Gutmann DH. Understanding a complicated gal-1[J]. Neuro-Oncology, 2019, 21(11):1341-1343.
doi: 10.1093/neuonc/noz165 pmid: 31538650 |
| [9] |
Shih TC, Liu R, Wu CT, et al. Targeting galectin-1 impairs castration-resistant prostate cancer progression and invasion[J]. Clin Cancer Res, 2018, 24(17):4319-4331.
doi: 10.1158/1078-0432.CCR-18-0157 URL |
| [10] |
Zhang P, Zhang P, Shi B, et al. Galectin-1 overexpression promotes progression and chemoresistance to cisplatin in epithelial ovarian cancer[J]. Cell Death Dis, 2014, 5(1):e991.
doi: 10.1038/cddis.2013.526 URL |
| [11] | Zhu J, Zheng Y, Zhang HY, et al. Galectin-1 induces metastasis and epithelial-mesenchymal transition(EMT) in human ovarian cancer cells via activation of the MAPK JNK/p38 signalling pathway[J]. Am J Transl Res, 2019, 11(6):3862-3878. |
| [12] |
Tang D, Yuan ZX, Xue XF, et al. High expression of Galectin-1 in pancreatic stellate cells plays a role in the development and maintenance of an immunosuppressive microenvironment in pancreatic cancer[J]. Int J Cancer, 2012, 130(10):2337-2348.
doi: 10.1002/ijc.26290 pmid: 21780106 |
| [13] |
Dalotto-Moreno T, Croci DO, Cerliani JP, et al. Targeting galectin-1 overcomes breast cancer-associated immunosuppression and prevents metastatic disease[J]. Cancer Res, 2013, 73(3):1107-1117.
doi: 10.1158/0008-5472.CAN-12-2418 pmid: 23204230 |
| [14] | Zhu X, Wang K, Zhang K, et al. Galectin-1 knockdown in carcinoma-associated fibroblasts inhibits migration and invasion of human MDA-MB-231 breast cancer cells by modulating MMP-9 expression[J]. Acta Biochim Biophys Sin(Shanghai), 2016, 48(5):462-467. |
| [15] |
Zhao HJ, Langerød A, Ji Y, et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast[J]. Mol Biol Cell, 2004, 15(6):2523-2536.
pmid: 15034139 |
| [16] |
Ito K, Park SH, Nayak A, et al. PTK6 inhibition suppresses metastases of triple-negative breast cancer via SNAIL-dependent E-cadherin regulation[J]. Cancer Res, 2016, 76(15):4406-4417.
doi: 10.1158/0008-5472.CAN-15-3445 pmid: 27302163 |
| [17] |
Huang YY, Wang HC, Zhao JW, et al. Immunosuppressive roles of galectin-1 in the tumor microenvironment[J]. Biomolecules, 2021, 11(10):1398.
doi: 10.3390/biom11101398 URL |
| [18] |
Perillo NL, Pace KE, Seilhamer JJ, et al. Apoptosis of T cells mediated by galectin-1[J]. Nature, 1995, 378(6558):736-739.
doi: 10.1038/378736a0 URL |
| [19] | 张丽娜, 杨艳芳, 姜战胜. PD-1/PD-L1抑制剂治疗三阴性乳腺癌的研究进展[J]. 中国肿瘤生物治疗杂志, 2021, 28(8):844-849. |
| Zhang LN, Yang YF, Jiang ZS. Research progress on PD-1/PD-L1 inhibitors in the treatment of triple-negative breast cancer[J]. Chin J Cancer Biotherapy, 2021, 28(8):844-849. |
| [1] | MA Yu-jing, DUAN Chun-hui, HE Ming-yang, ZHANG Ying-jie, YANG Ruo-chen, WANG Yong, LIU Yue-qin. Effects of Knockout of G0S2 Gene in Ovarian Granulosa Cell Proliferation, Steroids Hormones and Related Gene Expression [J]. Biotechnology Bulletin, 2023, 39(6): 325-334. |
| [2] | XV Ru-yue, WANG Zi-xiao, SHEN Lu, WU Rong-rong, YAO Fang-ting, TAN Zhong-yuan, LIU Heng-wei, ZHANG Wen-chao. Research Progress in Bioremediation of Cr(VI) [J]. Biotechnology Bulletin, 2023, 39(6): 49-60. |
| [3] | YANG Xiao-feng, QIN Xiao-wei, GUO Ze-yuan, LV Li-hua. Effects of Proanthocyanidin on the Proliferation of Sheep Follicular Granulosa Cells in vitro [J]. Biotechnology Bulletin, 2022, 38(9): 258-263. |
| [4] | YANG Xin-ran, WANG Jian-fang, MA Xin-hao, ZAN Lin-sen. Expression Analyses of m6A Methylase Genes in Bovine Adipogenesis [J]. Biotechnology Bulletin, 2022, 38(7): 70-79. |
| [5] | SHENG Xue-qing, ZHAO Nan, LIN Ya-qiu, CHEN Ding-shuang, WANG Rui-long, LI Ao, WANG Yong, LI Yan-yan. Cloning and Expression Analysis of ZNF32 Gene in Goat [J]. Biotechnology Bulletin, 2022, 38(12): 300-311. |
| [6] | JIN Qiu-xia, WANG Si-hong, JIN Li-hua. Research Progress on Drosophila Intestinal Stem Cells and Intestinal Microflora [J]. Biotechnology Bulletin, 2021, 37(4): 245-250. |
| [7] | ZHENG Fang-fang, LIN Jun-sheng. Selection and Specificity of Nucleic Acid Aptamers for a Proliferation Inducing Ligand [J]. Biotechnology Bulletin, 2021, 37(10): 196-202. |
| [8] | YANG Lei, YE Zhou-jie, LI Zhao-long, SHEN Yang-kun, FU Ya-juan. Effects of TET2 on T Cell Proliferation by Electroporation [J]. Biotechnology Bulletin, 2020, 36(1): 229-237. |
| [9] | LI Ping ,ZHANG Gui-ping, HU Jian-ran. Effects of Total Flavonoids from Forsythia suspense on the Proliferation of Gastric Cancer Cell MGC80-3 [J]. Biotechnology Bulletin, 2018, 34(6): 199-203. |
| [10] | DONG Ying, HU Hong-xia, TIAN Zhao-hui, WANG Wei, DONG Tian. Isolation of Peripheral Blood Lymphocytes of Sturgeon and the Optimal Proliferate Response Condition [J]. Biotechnology Bulletin, 2018, 34(3): 150-155. |
| [11] | YUAN Bai-yin, LIU Zhong-ying. Optimization of EDU Pulse Method for Tracing the Migration of Mouse Second Heart Field Cells [J]. Biotechnology Bulletin, 2018, 34(12): 84-89. |
| [12] | YUAN Bai-yin, HU Ji-sheng, LIU Zhong-ying, HUANG Xia. Wdr1 Knockout Inhibits Migration and Proliferation of Mouse Primary Vascular Smooth Muscle Cells [J]. Biotechnology Bulletin, 2018, 34(12): 179-185. |
| [13] | GUO Hong-yan, GAO Han, WU Qi, SUN Xiao-jie, LIU Xiu-cai, ZHAO Li-qun. Construction of SGK3 Gene Lentiviral RNA Interference Vector and Effects on Cell proliferation and Apoptosis of Breast Cancer Cell Line MB-474 [J]. Biotechnology Bulletin, 2018, 34(1): 247-252. |
| [14] | HU Jian-ran LI Ping LEI Hai-ying LIU Xian-rui. Effects of Polysaccharides from Lu Dangshen(Codonopsis pilosula)on Proliferation and Migration of Human Cervical SiHa [J]. Biotechnology Bulletin, 2017, 33(5): 159-163. |
| [15] | LIU Hong-bo, ZHENG Peng, YIN Ze, ZHANG Tong-cun. Knockdown of Long Non-coding RNA HOTAIR by CRISPR/Cas9 System [J]. Biotechnology Bulletin, 2017, 33(11): 180-187. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||