








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (1): 104-114.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0417
Previous Articles Next Articles
CHEN Quan-bing1( ), CAO Wei-jie1, LI Chun1,2, LV Bo1(
), CAO Wei-jie1, LI Chun1,2, LV Bo1( )
)
Received:2022-04-07
Online:2023-01-26
Published:2023-02-02
Contact:
LV Bo
E-mail:873569839@qq.com;lv-b@bit.edu.cn
CHEN Quan-bing, CAO Wei-jie, LI Chun, LV Bo. Molecular Evolutionary Relationship and Protein Structure of Glycoside Hydrolases from GH79 Family[J]. Biotechnology Bulletin, 2023, 39(1): 104-114.
| 水解方式 Hydrolysis method | 名称 Name | 切割位点 Cleavage site |
|---|---|---|
| 外切型 Exo-type | 唾液酸酶 | α-2,6糖苷键连接的唾液酸残基 |
| β-半乳糖苷酶 | β-1,4糖苷键连接的半乳糖残基 | |
| α-甘露糖苷酶 | α-1,2糖苷键,α-1,3糖苷键,α-1,6糖苷键连接的甘露糖型 | |
| β-甘露糖苷酶 | β-1,4糖苷键连接的甘露糖型 | |
| β-淀粉酶 | 自非还原末端依次切割麦芽糖单位,不切也不逾越α-1,6糖苷键 | |
| 内切型 Endo-type | α-淀粉酶 | 在分子内部随机切割α-1,4糖苷键,不切α-1,6糖苷键 |
| 异淀粉酶 | 水解支链淀粉或糖原中的α-1,6糖苷键 | |
| Endo F | β-1,4糖苷键连接的高甘露糖型杂合糖型 | |
| Endo H | β-1,4糖苷键连接的高甘露糖型杂合糖型 | |
| O-糖苷酶 | 水解蛋白核心1(Galβ1-3 GalNAC-Thr/Ser)和核心3(GlcNACβ1-3 GalNAC-Thr/Ser)之间的O-连接二糖单位 |
Table 1 Classification of glycoside hydrolases
| 水解方式 Hydrolysis method | 名称 Name | 切割位点 Cleavage site |
|---|---|---|
| 外切型 Exo-type | 唾液酸酶 | α-2,6糖苷键连接的唾液酸残基 |
| β-半乳糖苷酶 | β-1,4糖苷键连接的半乳糖残基 | |
| α-甘露糖苷酶 | α-1,2糖苷键,α-1,3糖苷键,α-1,6糖苷键连接的甘露糖型 | |
| β-甘露糖苷酶 | β-1,4糖苷键连接的甘露糖型 | |
| β-淀粉酶 | 自非还原末端依次切割麦芽糖单位,不切也不逾越α-1,6糖苷键 | |
| 内切型 Endo-type | α-淀粉酶 | 在分子内部随机切割α-1,4糖苷键,不切α-1,6糖苷键 |
| 异淀粉酶 | 水解支链淀粉或糖原中的α-1,6糖苷键 | |
| Endo F | β-1,4糖苷键连接的高甘露糖型杂合糖型 | |
| Endo H | β-1,4糖苷键连接的高甘露糖型杂合糖型 | |
| O-糖苷酶 | 水解蛋白核心1(Galβ1-3 GalNAC-Thr/Ser)和核心3(GlcNACβ1-3 GalNAC-Thr/Ser)之间的O-连接二糖单位 |
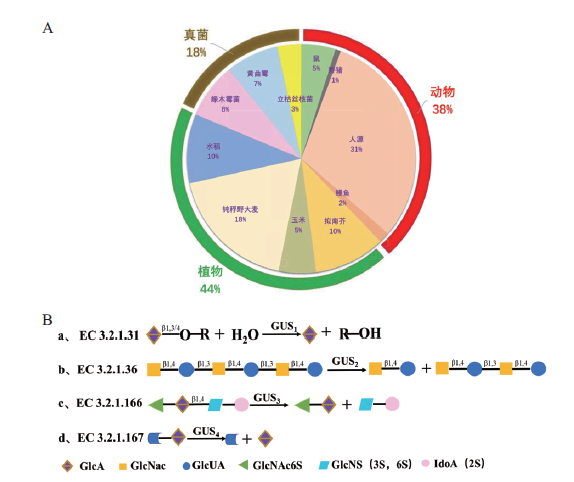
Fig. 1 Distribution and classification of catalytic types of glycoside hydrolases in the family GH79 A: Distribution of GH79 glycosidase from different sources. B: Molecular model of GH79 glycosidase catalysis
| 蛋白名称 Protein name | 来源 Organism | 底物 Substrate | EC编号 EC# | 参考文献 Reference |
|---|---|---|---|---|
| GlcA79A | Acidobacterium capsulatum | β-D-glucuronicacid | 3.2.1.31 | [ |
| AtGUS2 | Arabidopsis thaliana | pNPGlcA | 3.2.1.31 | [ |
| Nc6GAL | Neurospora crassa | Arabinogalactan-proteins | 3.2.1.31 | [ |
| FobglcA | Fusarium oxysporum | Gum arabic | 3.2.1.31 | [ |
| BpHep | Burkholderia pseudomallei | Heparan sulfate | 3.2.1.166 | [ |
| HpsE | Rattus norvegicus | Heparan sulfate | 3.2.1.166 | [ |
| Hpse1 | Danio rerio | Heparan sulfate glycosaminoglycans | 3.2.1.166 | [ |
| Hpa1 | Homo sapiens | Heparan sulfate | 3.2.1.166 | [ |
| Heparanase | Gallus gallus | Heparan sulfate | 3.2.1.166 | [ |
| Heparanase | Homo sapiens | Heparan sulfate | 3.2.1.166 | [ |
| Heparanase | Sus scrofa | Heparan sulfate | 3.2.1.166 | [ |
| Hpa | Mus musculus | Heparan sulfate | 3.2.1.166 | [ |
| Hyaluronidase | Hirudo nipponia | Hyaluronan | 3.2.1.36 | [ |
| LHyal | Hirudo nipponia | Hyaluronan | 3.2.1.36 | [ |
| SguS | Scutellaria baicalensis | Baicalein-7-O-β-D-glucuronide-conjugated | 3.2.1.167 | [ |
| AnGlcAase | Aspergillus niger | Arabinogalactan-proteins | 3.2.1.- | [ |
| TpGUS79A | Talaromyces pinophilus Li-93 | Glycyrrhizinate | 3.2.1.- | [ |
| Glucuronidase 1 | Prunus dulcis | Almond | 3.2.1.- | [ |
| Heparanase precursor | Bos taurus | Heparan sulfate | 3.2.1.- | [ |
| Hypothetical protein TrVGV298_008121 | Trichoderma virens | - | 3.2.1.- | [ |
Table 2 Different functional glycoside hydrolases of the GH79 family
| 蛋白名称 Protein name | 来源 Organism | 底物 Substrate | EC编号 EC# | 参考文献 Reference |
|---|---|---|---|---|
| GlcA79A | Acidobacterium capsulatum | β-D-glucuronicacid | 3.2.1.31 | [ |
| AtGUS2 | Arabidopsis thaliana | pNPGlcA | 3.2.1.31 | [ |
| Nc6GAL | Neurospora crassa | Arabinogalactan-proteins | 3.2.1.31 | [ |
| FobglcA | Fusarium oxysporum | Gum arabic | 3.2.1.31 | [ |
| BpHep | Burkholderia pseudomallei | Heparan sulfate | 3.2.1.166 | [ |
| HpsE | Rattus norvegicus | Heparan sulfate | 3.2.1.166 | [ |
| Hpse1 | Danio rerio | Heparan sulfate glycosaminoglycans | 3.2.1.166 | [ |
| Hpa1 | Homo sapiens | Heparan sulfate | 3.2.1.166 | [ |
| Heparanase | Gallus gallus | Heparan sulfate | 3.2.1.166 | [ |
| Heparanase | Homo sapiens | Heparan sulfate | 3.2.1.166 | [ |
| Heparanase | Sus scrofa | Heparan sulfate | 3.2.1.166 | [ |
| Hpa | Mus musculus | Heparan sulfate | 3.2.1.166 | [ |
| Hyaluronidase | Hirudo nipponia | Hyaluronan | 3.2.1.36 | [ |
| LHyal | Hirudo nipponia | Hyaluronan | 3.2.1.36 | [ |
| SguS | Scutellaria baicalensis | Baicalein-7-O-β-D-glucuronide-conjugated | 3.2.1.167 | [ |
| AnGlcAase | Aspergillus niger | Arabinogalactan-proteins | 3.2.1.- | [ |
| TpGUS79A | Talaromyces pinophilus Li-93 | Glycyrrhizinate | 3.2.1.- | [ |
| Glucuronidase 1 | Prunus dulcis | Almond | 3.2.1.- | [ |
| Heparanase precursor | Bos taurus | Heparan sulfate | 3.2.1.- | [ |
| Hypothetical protein TrVGV298_008121 | Trichoderma virens | - | 3.2.1.- | [ |
| [1] |
OKUYAMA M. Function and structure studies of GH family 31 and 97 α-glycosidases[J]. Biosci Biotechnol Biochem, 2011, 75(12): 2269-2277.
doi: 10.1271/bbb.110610 URL |
| [2] |
Larrucea S, Butta N, Arias-Salgado EG, et al. Expression of podocalyxin enhances the adherence, migration, and intercellular communication of cells[J]. Exp Cell Res, 2008, 314(10): 2004-2015.
doi: 10.1016/j.yexcr.2008.03.009 pmid: 18456258 |
| [3] |
Singh RS, Singh RP, Kennedy JF. Recent insights in enzymatic synthesis of fructooligosaccharides from inulin[J]. Int J Biol Macromol, 2016, 85:565-572.
doi: 10.1016/j.ijbiomac.2016.01.026 pmid: 26791586 |
| [4] |
Guo LC, Katiyo W, Lu LS, et al. Glycyrrhetic acid 3-O-mono-β-d-glucuronide(GAMG):an innovative high-potency sweetener with improved biological activities[J]. Compr Rev Food Sci Food Saf, 2018, 17(4): 905-919.
doi: 10.1111/1541-4337.12353 URL |
| [5] | Lu DQ, Zhang SM, Wang J, et al. Adsorption separation of 3 beta-D-monoglucuronyl-18 beta-glycyrrhetinic acid from directional biotransformation products of glycyrrhizin[J]. African J Biotechnol, 2008, 7(22): 3995-4003. |
| [6] |
Tang WJ, Yang YA, Xu H, et al. Synthesis and discovery of 18α-GAMG as anticancer agent in vitro and in vivo via down expression of protein p65[J]. Sci Rep, 2014, 4:7106.
doi: 10.1038/srep07106 URL |
| [7] |
Mitsudome T, Xu J, Nagata Y, et al. Expression, purification, and characterization of endo-β-N-acetylglucosaminidase H using baculovirus-mediated silkworm protein expression system[J]. Appl Biochem Biotechnol, 2014, 172(8): 3978-3988.
doi: 10.1007/s12010-014-0814-5 pmid: 24599668 |
| [8] |
Komba S, Ito Y. Β-galactosidase-catalyzed intramolecular transglycosylation[J]. Tetrahedron Lett, 2001, 42(48): 8501-8505.
doi: 10.1016/S0040-4039(01)01826-3 URL |
| [9] |
Davies GJ, Williams SJ. Carbohydrate-active enzymes:sequences, shapes, contortions and cells[J]. Biochem Soc Trans, 2016, 44(1): 79-87.
doi: 10.1042/BST20150186 URL |
| [10] |
Niers TMH, Klerk CPW, DiNisio M, et al. Mechanisms of heparin induced anti-cancer activity in experimental cancer models[J]. Crit Rev Oncol Hematol, 2007, 61(3): 195-207.
doi: 10.1016/j.critrevonc.2006.07.007 URL |
| [11] |
Hamre AG, Strømnes AGS, Gustavsen D, et al. Treatment of recalcitrant crystalline polysaccharides with lytic polysaccharide monooxygenase relieves the need for glycoside hydrolase processivity[J]. Carbohydr Res, 2019, 473:66-71.
doi: 10.1016/j.carres.2019.01.001 URL |
| [12] |
Saito A, Wakao M, Deguchi H, et al. Towards the assembly of heparin and heparan sulfate oligosaccharide libraries:efficient synthesis of uronic acid and disaccharide building blocks[J]. Tetrahedron, 2010, 66(22): 3951-3962.
doi: 10.1016/j.tet.2010.03.077 URL |
| [13] |
Michikawa M, Ichinose H, Momma M, et al. Structural and biochemical characterization of glycoside hydrolase family 79 β-glucuronidase from Acidobacterium capsulatum[J]. J Biol Chem, 2012, 287(17): 14069-14077.
doi: 10.1074/jbc.M112.346288 pmid: 22367201 |
| [14] |
Bohlmann L, Tredwell GD, Yu X, et al. Functional and structural characterization of a heparanase[J]. Nat Chem Biol, 2015, 11(12): 955-957.
doi: 10.1038/nchembio.1956 pmid: 26565989 |
| [15] |
Konishi T, Kotake T, Soraya D, et al. Properties of family 79 beta-glucuronidases that hydrolyze beta-glucuronosyl and 4-O-methyl-beta-glucuronosyl residues of arabinogalactan-protein[J]. Carbohydr Res, 2008, 343(7): 1191-1201.
doi: 10.1016/j.carres.2008.03.004 URL |
| [16] |
Eudes A, Mouille G, Thévenin J, et al. Purification, cloning and functional characterization of an endogenous beta-glucuronidase in Arabidopsis thaliana[J]. Plant Cell Physiol, 2008, 49(9): 1331-1341.
doi: 10.1093/pcp/pcn108 URL |
| [17] |
Hulett MD, Freeman C, Hamdorf BJ, et al. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis[J]. Nat Med, 1999, 5(7): 803-809.
doi: 10.1038/10525 pmid: 10395326 |
| [18] |
Podyma-Inoue KA, Yokote H, Sakaguchi K, et al. Characterization of heparanase from a rat parathyroid cell line[J]. J Biol Chem, 2002, 277(36): 32459-32465.
doi: 10.1074/jbc.M203282200 pmid: 12077130 |
| [19] |
Toyoshima M, Nakajima M. Human heparanase. purification, characterization, cloning, and expression[J]. J Biol Chem, 1999, 274(34): 24153-24160.
doi: 10.1074/jbc.274.34.24153 pmid: 10446189 |
| [20] |
Masola V, Bellin G, Gambaro G, et al. Heparanase:a multitasking protein involved in extracellular matrix(ECM)remodeling and intracellular events[J]. Cells, 2018, 7(12): 236.
doi: 10.3390/cells7120236 URL |
| [21] |
Kondo T, Kichijo M, Nakaya M, et al. Biochemical and structural characterization of a novel 4-O-α-l-rhamnosyl-β-d-glucuronidase from Fusarium oxysporum[J]. FEBS J, 2021, 288(16): 4918-4938.
doi: 10.1111/febs.15795 URL |
| [22] |
Wei KH, Liu IH. Heparan sulfate glycosaminoglycans modulate migration and survival in zebrafish primordial germ cells[J]. Theriogenology, 2014, 81(9): 1275-1285. e1-2.
doi: 10.1016/j.theriogenology.2014.02.009 URL |
| [23] |
Goldshmidt O, Zcharia E, Aingorn H, et al. Expression pattern and secretion of human and chicken heparanase are determined by their signal peptide sequence[J]. J Biol Chem, 2001, 276(31): 29178-29187.
doi: 10.1074/jbc.M102462200 pmid: 11387326 |
| [24] |
Strausberg RL, Feingold EA, Grouse LH, et al. Generation and initial analysis of more than 15, 000 full-length human and mouse cDNA sequences[J]. Proc Natl Acad Sci USA, 2002, 99(26): 16899-16903.
doi: 10.1073/pnas.242603899 pmid: 12477932 |
| [25] |
Miles JR, Vallet JL, Freking BA, et al. Molecular cloning and characterisation of heparanase mRNA in the porcine placenta throughout gestation[J]. Reprod Fertil Dev, 2009, 21(6): 757-772.
doi: 10.1071/RD09041 URL |
| [26] |
Jin P, Kang Z, Zhang N, et al. High-yield novel leech hyaluronidase to expedite the preparation of specific hyaluronan oligomers[J]. Sci Rep, 2014, 4:4471.
doi: 10.1038/srep04471 pmid: 24667183 |
| [27] | Huang H, Hou XD, Xu RR, et al. Structure and cleavage pattern of a hyaluronate 3-glycanohydrolase in the glycoside hydrolase 79 family[J]. Carbohydr Polym, 2022, 277:118838. |
| [28] |
Sasaki K, Taura F, Shoyama Y, et al. Molecular characterization of a novel beta-glucuronidase from Scutellaria baicalensis Georgi[J]. J Biol Chem, 2000, 275(35): 27466-27472.
doi: 10.1074/jbc.M004674200 pmid: 10858442 |
| [29] | Xu YH, Feng XD, Jia JT, et al. A novel β-glucuronidase from Talaromyces pinophilus Li-93 precisely hydrolyzes glycyrrhizin into glycyrrhetinic acid 3- O-mono-β- d-glucuronide[J]. Appl Environ Microbiol, 2018, 84(19): e00755-18. |
| [30] |
Sánchez-Pérez R, Pavan S, Mazzeo R, et al. Mutation of a bHLH transcription factor allowed almond domestication[J]. Science, 2019, 364(6445): 1095-1098.
doi: 10.1126/science.aav8197 pmid: 31197015 |
| [31] |
Hambruch N, Kumstel S, Haeger JD, et al. Bovine placentomal heparanase and syndecan expression is related to placental maturation[J]. Placenta, 2017, 57:42-51.
doi: S0143-4004(17)30290-4 pmid: 28864018 |
| [32] | Li WC, Lin TC, Chen CL, et al. Complete genome sequences and genome-wide characterization of Trichoderma biocontrol agents provide new insights into their evolution and variation in genome organization, sexual development, and fungal-plant interactions[J]. Microbiol Spectr, 2021, 9(3): e0066321. |
| [33] |
Wu L, Jiang JB, Jin Y, et al. Activity-based probes for functional interrogation of retaining β-glucuronidases[J]. Nat Chem Biol, 2017, 13(8): 867-873.
doi: 10.1038/nchembio.2395 pmid: 28581485 |
| [34] |
Li QF, Jiang T, Liu R, et al. Tuning the pH profile of β-glucuronidase by rational site-directed mutagenesis for efficient transformation of glycyrrhizin[J]. Appl Microbiol Biotechnol, 2019, 103(12): 4813-4823.
doi: 10.1007/s00253-019-09790-3 pmid: 31055652 |
| [35] |
Vlodavsky I, Ilan N, Naggi A, et al. Heparanase:structure, biological functions, and inhibition by heparin-derived mimetics of heparan sulfate[J]. Curr Pharm Des, 2007, 13(20): 2057-2073.
doi: 10.2174/138161207781039742 URL |
| [36] |
Wang F, Wan A, Rodrigues B. The function of heparanase in diabetes and its complications[J]. Can J Diabetes, 2013, 37(5): 332-338.
doi: 10.1016/j.jcjd.2013.05.008 pmid: 24500561 |
| [37] |
Kuroyama H, Tsutsui N, Hashimoto Y, et al. Purification and characterization of a beta-glucuronidase from Aspergillus niger[J]. Carbohydr Res, 2001, 333(1): 27-39.
doi: 10.1016/S0008-6215(01)00114-8 URL |
| [38] | Harty DWS, Mayo JA, Cook SL, et al. Environmental regulation of glycosidase and peptidase production by Streptococcus gordonii FSS2[J]. Microbiology(Reading), 2000, 146(Pt 8): 1923-1931. |
| [39] |
Imaki H, Tomoyasu T, Yamamoto N, et al. Identification and characterization of a novel secreted glycosidase with multiple glycosidase activities in Streptococcus intermedius[J]. J Bacteriol, 2014, 196(15): 2817-2826.
doi: 10.1128/JB.01727-14 URL |
| [40] | Tomoyasu T, Yamasaki T, Chiba S, et al. Positive- and negative-control pathways by blood components for intermedilysin production in Streptococcus intermedius[J]. Infect Immun, 2017, 85(9): e00379-e00317. |
| [41] |
Fratianni F, Ombra MN, D'Acierno A, et al. Polyphenols content and in vitro α-glycosidase activity of different Italian monofloral honeys, and their effect on selected pathogenic and probiotic bacteria[J]. Microorganisms, 2021, 9(8): 1694.
doi: 10.3390/microorganisms9081694 URL |
| [42] | Weber BA, Klein JR, Henrich B. Expression of the phospho-beta-glycosidase ArbZ from Lactobacillus delbrueckii subsp. lactis in Lactobacillus helveticus:substrate induction and catabolite repression[J]. Microbiology(Reading), 2000, 146(Pt 8): 1941-1948. |
| [43] |
Chang CF, Ho CW, Wu CY, et al. Discovery of picomolar slow tight-binding inhibitors of alpha-fucosidase[J]. Chem Biol, 2004, 11(9): 1301-1306.
doi: 10.1016/j.chembiol.2004.07.009 URL |
| [44] |
Liu L, Tharmalingam T, Maischberger E, et al. A HPLC-based glycoanalytical protocol allows the use of natural O-glycans derived from glycoproteins as substrates for glycosidase discovery from microbial culture[J]. Glycoconj J, 2013, 30(8): 791-800.
doi: 10.1007/s10719-013-9483-9 URL |
| [45] |
Zhang LW, Qiu HL, Yuan S, et al. Revelation of mechanism for aqueous saponins content decrease during storage of Dioscorea zingiberensis C. H. Wright tubers:an essential prerequisite to ensure clean production of diosgenin[J]. Ind Crops Prod, 2018, 125:178-185.
doi: 10.1016/j.indcrop.2018.09.003 URL |
| [46] |
Brooks JF II, Gyllborg MC, Cronin DC, et al. Global discovery of colonization determinants in the squid symbiont Vibrio fischeri[J]. PNAS, 2014, 111(48): 17284-17289.
doi: 10.1073/pnas.1415957111 pmid: 25404340 |
| [47] | Zhang BQ, Zhang N, Zhang QQ, et al. Transcriptome profiles of Sporisorium reilianum during the early infection of resistant and susceptible maize isogenic lines[J]. J Fungi(Basel), 2021, 7(2): 150. |
| [48] |
Novakazi F, Göransson M, Stefánsson TS, et al. Virulence of Icelandic Pyrenophora teres f. teres populations and resistance of Icelandic spring barley lines[J]. J Plant Pathol, 2022, 104(1): 205-213.
doi: 10.1007/s42161-021-00972-5 URL |
| [49] |
Hall BG. Building phylogenetic trees from molecular data with MEGA[J]. Mol Biol Evol, 2013, 30(5): 1229-1235.
doi: 10.1093/molbev/mst012 pmid: 23486614 |
| [50] |
Newman L, Duffus ALJ, Lee C. Using the free program MEGA to build phylogenetic trees from molecular data[J]. Am Biol Teach, 2016, 78(7): 608-612.
doi: 10.1525/abt.2016.78.7.608 URL |
| [51] |
Vinader V, Haji-Abdullahi MH, Patterson LH, et al. Synthesis of a pseudo-disaccharide library and its application to the characterisation of the heparanase catalytic site[J]. PLoS One, 2013, 8(11): e82111.
doi: 10.1371/journal.pone.0082111 URL |
| [52] |
Okuyama M, Yoshida T, Hondoh H, et al. Catalytic role of the calcium ion in GH97 inverting glycoside hydrolase[J]. FEBS Lett, 2014, 588(17): 3213-3217.
doi: 10.1016/j.febslet.2014.07.002 pmid: 25017438 |
| [53] |
Qin Z, Yang SQ, Zhao LM, et al. Catalytic mechanism of a novel glycoside hydrolase family 16 “elongating” β-transglycosylase[J]. J Biol Chem, 2017, 292(5): 1666-1678.
doi: 10.1074/jbc.M116.762419 URL |
| [54] |
Vuong TV, Wilson DB. Glycoside hydrolases:catalytic base/nucleophile diversity[J]. Biotechnol Bioeng, 2010, 107(2): 195-205.
doi: 10.1002/bit.22838 pmid: 20552664 |
| [55] |
Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases[J]. Structure, 1995, 3(9): 853-859.
doi: 10.1016/S0969-2126(01)00220-9 pmid: 8535779 |
| [56] |
Wu L, Viola CM, Brzozowski AM, et al. Structural characterization of human heparanase reveals insights into substrate recognition[J]. Nat Struct Mol Biol, 2015, 22(12): 1016-1022.
doi: 10.1038/nsmb.3136 pmid: 26575439 |
| [1] | ZHAO Yu-xue, WANG Yun, YU Lu-yao, LIU Jing-jing, SI Jin-ping, ZHANG Xin-feng, ZHANG Lei. Structure and Application of C-glycosyltransferases in Plants [J]. Biotechnology Bulletin, 2022, 38(10): 18-28. |
| [2] | CHEN Ming-yu, NI Xuan, SI You-bin, SUN Kai. Advances in the Application of Immobilized Fungal Laccase for the Bioremediation of Environmental Organic Contamination [J]. Biotechnology Bulletin, 2021, 37(6): 244-258. |
| [3] | BAI Fu-mei, LI Zhi-min, WANG Xiao-qin, HU Zi-wei, BAO Ling-ling, LI Zhi-min. Biochemical Characterization and Structural Analysis of N-acetylornithine Transaminase from Synechocystis sp. PCC6803 [J]. Biotechnology Bulletin, 2021, 37(5): 98-107. |
| [4] | SHI Li-xia, GAO Song-feng, ZHU Lei-lei. Research Advance in Polyethylene Terephthalate Hydrolytic Enzymes [J]. Biotechnology Bulletin, 2020, 36(10): 226-236. |
| [5] | QIAO Jing, CUI Sheng-rong, SHI Hong-wu, LUO Zu-liang, MA Xiao-jun. Homology Modeling and Molecular Docking of Cycloartenol Synthase in Siraitia grosvenorii and Speculated Mechanism of Catalytic Cyclization [J]. Biotechnology Bulletin, 2019, 35(2): 101-108. |
| [6] | LI Bin,CHEN Xiang-nan,ZHANG Jian-fa,WANG Shi-ming. Screening of Exopolysaccharide-producing Strains and Structural Analysis of the Exopolysaccharides [J]. Biotechnology Bulletin, 2016, 32(5): 165-171. |
| [7] | Jiang Ping, Erdemtu, Gao Fengshan. The Cloning of cDNA of SLA-DRB from Hebao Pigs and the Analysis of Their Molecular Evolutionary Characteristics [J]. Biotechnology Bulletin, 2015, 31(12): 150-157. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||