








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (1): 115-126.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0378
Previous Articles Next Articles
ZHOU Jia-yan1( ), ZOU Jian1, CHEN Wei-ying1, WU Yi-chao1, CHEN Xi-tong1, WANG Qian1, ZENG Wen-jing1, HU Nan2(
), ZOU Jian1, CHEN Wei-ying1, WU Yi-chao1, CHEN Xi-tong1, WANG Qian1, ZENG Wen-jing1, HU Nan2( ), YANG Jun1(
), YANG Jun1( )
)
Received:2022-03-29
Online:2023-01-26
Published:2023-02-02
Contact:
HU Nan,YANG Jun
E-mail:601163528@qq.com;tomatohu2018@163.com;yangjun@cwnu.edu.cn
ZHOU Jia-yan, ZOU Jian, CHEN Wei-ying, WU Yi-chao, CHEN Xi-tong, WANG Qian, ZENG Wen-jing, HU Nan, YANG Jun. Construction of Multi-gene Interference System for Plant and Analysis of Its Application Efficiency[J]. Biotechnology Bulletin, 2023, 39(1): 115-126.
| 基因名称 Gene name | 番茄数据库基因ID Gene ID in tomato genome database |
|---|---|
| SlPP2C1 | Solyc03g121880.2.1 |
| SlPP2C2 | Solyc12g096020.1.1 |
| SlPP2C3 SlPP2C4 | Solyc08g062650.2.1 Solyc07g040990.2.1 |
Table 1 Information of four PP2C genes used in this paper
| 基因名称 Gene name | 番茄数据库基因ID Gene ID in tomato genome database |
|---|---|
| SlPP2C1 | Solyc03g121880.2.1 |
| SlPP2C2 | Solyc12g096020.1.1 |
| SlPP2C3 SlPP2C4 | Solyc08g062650.2.1 Solyc07g040990.2.1 |
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Purpose |
|---|---|---|
| SlPP2C1-F正 | GCGAGCTCGTTGATGTTGGTAGAGTTCCCTTG | 干扰片段扩增引物 Primers for amplifying interference fragment |
| SlPP2C1-R-B | TCCGAATCAGCCTTTCTATCTAAACTCCTTTCTTCC | |
| SlPP2C2-F-B | GATAGAAAGGCTGATTCGGAGAGTGACCTTAG | |
| SlPP2C2-R正 | ACGCGTCGACGAGAATCCACGAAATCCTGACC | |
| SlPP2C1-F反 | CTCTAGAGTTGATGTTGGTAGAGTTCCCTTG | |
| SlPP2C1-R-B | TCCGAATCAGCCTTTCTATCTAAACTCCTTTCTTCC | |
| SlPP2C2-F-B | GATAGAAAGGCTGATTCGGAGAGTGACCTTAG | |
| SlPP2C2-R反 | TCCCCCGGGGAGAATCCACGAAATCCTGACC | |
| SlPP2C3-F正 | GCGAGCTCATGTGTTTGGCGGTTGCTTTGC | |
| SlPP2C3-R-B | CTATGATTCGGCATCTACCAAATTCTGTCCGTC | |
| SlPP2C4-F-B | TGGTAGATGCCGAATCATAGAGTGTCCAGGG | |
| SlPP2C4-R正 | ACGCGTCGACTTAGCCAAACTACACGAAAAAGAC | |
| SlPP2C3-F反 | CTCTAGAATGTGTTTGGCGGTTGCTTTGC | |
| SlPP2C3-R-B | CTATGATTCGGCATCTACCAAATTCTGTCCGTC | |
| SlPP2C4-F-B | TGGTAGATGCCGAATCATAGAGTGTCCAGGG | |
| SlPP2C4-R反 | TCCCCCGGGTTAGCCAAACTACACGAAAAAGAC | |
| qSlPP2C1-F | TAGCTGCACCTCTGAGCCTA | RT-qPCR引物Primers for RT-qPCR |
| qSlPP2C1-R | CTGCTTCTTTGCCACGATAA | |
| qSlPP2C2-F | CAGCAGAATGCTTGTCGAAT | |
| qSlPP2C2-R | ATGAGGCCAATTGTGTTGAA | |
| qSlPP2C3-F | GTCGCCATTGTTTGTTCATC | |
| qSlPP2C3-R | TCTTCTCGATTTGGCTTGTG | |
| qSlPP2C4-F | GATGGGCTATGGGATGTCTT | |
| qSlPP2C4-R | CTTGAGCAGCAGGATCTACG | |
| qSlUBI-F | GCCGACTACAACATCCAGAAGG | RT-qPCR内参引物Reference primers for RT-qPCR |
| qSlUBI-R | TGCAACACAGCGAGCTTAACC | |
| Loop-JC-F | ACAAGTTCAGCGTGTCCGGCGA | 载体元件特异引物Special primers for vector element |
| Loop-JC-R | TCGCCGGACACGCTGAACTTGT |
Table 2 All primers used in this study
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Purpose |
|---|---|---|
| SlPP2C1-F正 | GCGAGCTCGTTGATGTTGGTAGAGTTCCCTTG | 干扰片段扩增引物 Primers for amplifying interference fragment |
| SlPP2C1-R-B | TCCGAATCAGCCTTTCTATCTAAACTCCTTTCTTCC | |
| SlPP2C2-F-B | GATAGAAAGGCTGATTCGGAGAGTGACCTTAG | |
| SlPP2C2-R正 | ACGCGTCGACGAGAATCCACGAAATCCTGACC | |
| SlPP2C1-F反 | CTCTAGAGTTGATGTTGGTAGAGTTCCCTTG | |
| SlPP2C1-R-B | TCCGAATCAGCCTTTCTATCTAAACTCCTTTCTTCC | |
| SlPP2C2-F-B | GATAGAAAGGCTGATTCGGAGAGTGACCTTAG | |
| SlPP2C2-R反 | TCCCCCGGGGAGAATCCACGAAATCCTGACC | |
| SlPP2C3-F正 | GCGAGCTCATGTGTTTGGCGGTTGCTTTGC | |
| SlPP2C3-R-B | CTATGATTCGGCATCTACCAAATTCTGTCCGTC | |
| SlPP2C4-F-B | TGGTAGATGCCGAATCATAGAGTGTCCAGGG | |
| SlPP2C4-R正 | ACGCGTCGACTTAGCCAAACTACACGAAAAAGAC | |
| SlPP2C3-F反 | CTCTAGAATGTGTTTGGCGGTTGCTTTGC | |
| SlPP2C3-R-B | CTATGATTCGGCATCTACCAAATTCTGTCCGTC | |
| SlPP2C4-F-B | TGGTAGATGCCGAATCATAGAGTGTCCAGGG | |
| SlPP2C4-R反 | TCCCCCGGGTTAGCCAAACTACACGAAAAAGAC | |
| qSlPP2C1-F | TAGCTGCACCTCTGAGCCTA | RT-qPCR引物Primers for RT-qPCR |
| qSlPP2C1-R | CTGCTTCTTTGCCACGATAA | |
| qSlPP2C2-F | CAGCAGAATGCTTGTCGAAT | |
| qSlPP2C2-R | ATGAGGCCAATTGTGTTGAA | |
| qSlPP2C3-F | GTCGCCATTGTTTGTTCATC | |
| qSlPP2C3-R | TCTTCTCGATTTGGCTTGTG | |
| qSlPP2C4-F | GATGGGCTATGGGATGTCTT | |
| qSlPP2C4-R | CTTGAGCAGCAGGATCTACG | |
| qSlUBI-F | GCCGACTACAACATCCAGAAGG | RT-qPCR内参引物Reference primers for RT-qPCR |
| qSlUBI-R | TGCAACACAGCGAGCTTAACC | |
| Loop-JC-F | ACAAGTTCAGCGTGTCCGGCGA | 载体元件特异引物Special primers for vector element |
| Loop-JC-R | TCGCCGGACACGCTGAACTTGT |

Fig. 1 Multi-genes silencing system A:Original vector system pCAMBIA1301. B:Modified vector system pCAMBIA1301m. C:Modified vector system pCAMBIA1301s. D:Schematic diagram of multi-genes interference vector system

Fig 3 Construction of SlPP2 Cs silencing vector and detection of positive plants A:Schematic diagram of SlPP2Cs multi-genes interference. B:GUS stain. WT:Wild-type tomato,T0-1,T0-2,T0-3 and T0-4 representing transgenic tomato. C:Direct PCR electrophoresis. M:2 kb DNA marker,1-5:showing reverse fragment of SlPP2C1,6-10:showing forward fragment of SlPP2C1;11-15:showing reverse fragment of SlPP2C2;16-20:showing forward fragment of SlPP2C2;21-25:showing reverse fragment of SlPP2C3;26-30:showing forward fragment of SlPP2C3;31-35 showing reverse fragment of SlPP2C4;36-40:showing forward fragment of SlPP2C4. 1-40 is the cyclic arrangement of WT,T0-1,T0-2,T0-3 and T0-4
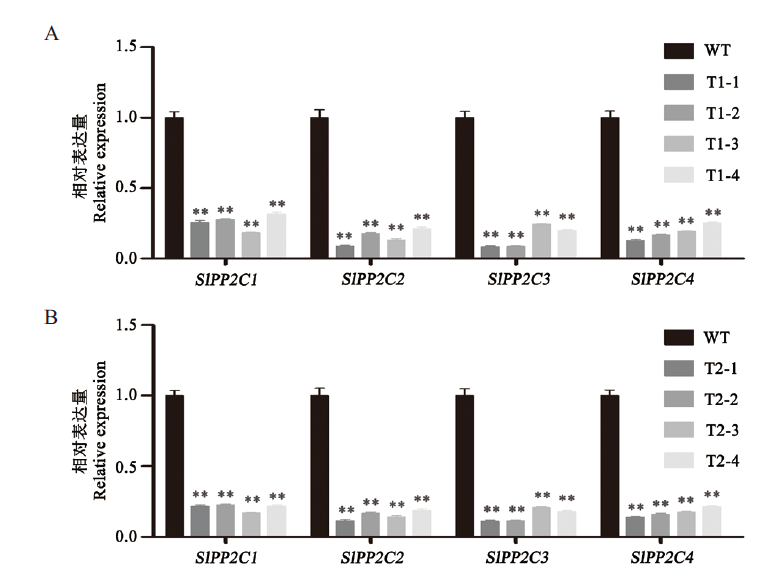
Fig. 4 Expression changes of four target genes after interf-erence A:Expression changes of four target genes in T1 generation transgenic plants. B:Expression changes of four target genes in T2 generation transgenic plants. Student's t test analysis,*:Compared with WT,P<0.05. **:Compared with WT,P<0.01. WT:Wild-type,T1-1,T1-2,T1-3,T1-4,T2-1,T2-2,T2-3 and T2-4 representing transgenic tomato. The same below
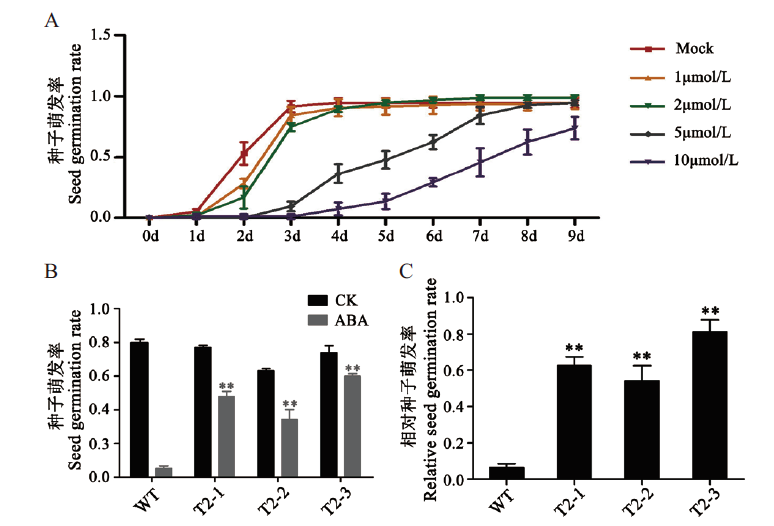
Fig. 5 Sensitivity test of transgenic RNAi plants to exoge-nous ABA A:Sensitivity of WT to exogenous ABA. X-axis represent time,9 d in total. B:Seed germination rate of interferred plants. CK:Double distilled water treatment. C:Relative seed germination rate of interfering plants
| [1] | Jack T. Molecular and genetic mechanisms of floral control[J]. Plant Cell, 2004, 16(Suppl):S1-S17. |
| [2] |
Ditta G, Pinyopich A, Robles P, et al. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity[J]. Curr Biol, 2004, 14(21): 1935-1940.
doi: 10.1016/j.cub.2004.10.028 pmid: 15530395 |
| [3] |
Pelaz S, Ditta GS, Baumann E, et al. B and C floral organ identity functions require SEPALLATA MADS-box genes[J]. Nature, 2000, 405(6783): 200-203.
doi: 10.1038/35012103 URL |
| [4] |
Yoshida T, Nishimura N, Kitahata N, et al. ABA-hypersensitive germination3 encodes a protein phosphatase 2C(AtPP2CA)that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs[J]. Plant Physiol, 2006, 140(1): 115-126.
pmid: 16339800 |
| [5] |
Leymarie J, Vavasseur A, Lascève G. CO2 sensing in stomata of abi1-1 and abi2-1 mutants of Arabidopsis thaliana[J]. Plant Physiol Biochem, 1998, 36(7): 539-543.
doi: 10.1016/S0981-9428(98)80180-0 URL |
| [6] |
Win J, Morgan W, Bos J, et al. Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes[J]. Plant Cell, 2007, 19(8): 2349-2369.
doi: 10.1105/tpc.107.051037 pmid: 17675403 |
| [7] | 韩长志. 植物病原卵菌RxLR效应基因功能研究进展[J]. 北方园艺, 2014(5): 188-193. |
| Han CZ. Research advance on functional effect of gene plant pathogenic oomycete[J]. North Hortic, 2014(5): 188-193. | |
| [8] |
Fujita Y, Fujita M, Shinozaki K, et al. ABA-mediated transcriptional regulation in response to osmotic stress in plants[J]. J Plant Res, 2011, 124(4): 509-525.
doi: 10.1007/s10265-011-0412-3 pmid: 21416314 |
| [9] | 胡楠. 快速易用开放的植物CRISPR/Cas9系统研发[D]. 重庆: 重庆大学, 2018. |
| Hu N. Development of rapid and user-friendly open-source CRISPR/Cas9 system[D]. Chongqing: Chongqing University, 2018. | |
| [10] |
Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells[J]. Science, 2002, 296(5567): 550-553.
doi: 10.1126/science.1068999 pmid: 11910072 |
| [11] |
Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs[J]. Genes Dev, 2001, 15(2): 188-200.
doi: 10.1101/gad.862301 URL |
| [12] |
Kerschen A, Napoli CA, Jorgensen RA, et al. Effectiveness of RNA interference in transgenic plants[J]. FEBS Lett, 2004, 566(1/2/3): 223-228.
doi: 10.1016/j.febslet.2004.04.043 URL |
| [13] |
Wesley SV, Helliwell CA, Smith NA, et al. Construct design for efficient, effective and high-throughput gene silencing in plants[J]. Plant J, 2001, 27(6): 581-590.
doi: 10.1046/j.1365-313x.2001.01105.x pmid: 11576441 |
| [14] |
Dalmay T, Hamilton A, Rudd S, et al. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus[J]. Cell, 2000, 101(5): 543-553.
doi: 10.1016/s0092-8674(00)80864-8 pmid: 10850496 |
| [15] | 徐进. 马铃薯StSUT2基因干扰载体的构建及遗传转化[D]. 兰州: 兰州理工大学, 2017. |
| Xu J. Construction of RNA interference vector of potato sucrose transporter 2 and its genetic transformation[D]. Lanzhou: Lanzhou University of Technology, 2017. | |
| [16] | 王锦达. 赤拟谷盗RNAi及dsRNA脱靶效应的研究[D]. 南京: 南京农业大学, 2015. |
| Wang JD. RNAi in Tribolium castaneum and the off target effect of dsRNA[D]. Nanjing: Nanjing Agricultural University, 2015. | |
| [17] |
Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi[J]. Nat Biotechnol, 2003, 21(6): 635-637.
doi: 10.1038/nbt831 pmid: 12754523 |
| [18] | 栾颖, 梁晋刚, 周晓莉, 等. RNAi转基因作物安全评价研究进展[J]. 生物安全学报, 2019, 28(2): 95-102. |
| Luan Y, Liang JG, Zhou XL, et al. Discussion on safety evaluation of RNAi transgenic crops[J]. J Biosaf, 2019, 28(2): 95-102. | |
| [19] |
Zhang GF, Zhao HT, Zhang CG, et al. TCP7 functions redundantly with several Class I TCPs and regulates endoreplication in Arabidopsis[J]. J Integr Plant Biol, 2019, 61(11): 1151-1170.
doi: 10.1111/jipb.12749 URL |
| [20] | 武娟. PAD4参与MPK4调控的拟南芥生长发育机制研究[D]. 北京: 中国农业大学, 2017. |
| Wu J. The regulatory mechanism of PAD4 in MPK4-regulated plant growth and development in Arabidopsis[D]. Beijing: China Agricultural University, 2017. | |
| [21] | 徐雪珍, 郑月萍, 张夏婷, 等. 拟南芥AtFAD6基因突变体的构建[J]. 江苏农业学报, 2021, 37(5): 1125-1130. |
| Xu XZ, Zheng YP, Zhang XT, et al. Construction of Arabidopsis AtFAD6 gene mutant[J]. Jiangsu J Agric Sci, 2021, 37(5): 1125-1130. | |
| [22] | 柴国华, 白泽涛, 蔡丽, 等. 油菜基因BnWRI1的克隆及RNAi对种子含油量的影响[J]. 中国农业科学, 2009, 42(5): 1512-1518. |
| Chai GH, Bai ZT, Cai L, et al. Cloning of BnWRI1 gene and the effect of RNA interference on seed oil content in oilseed rape[J]. Sci Agric Sin, 2009, 42(5): 1512-1518. | |
| [23] |
Peng Q, Hu Y, Wei R, et al. Simultaneous silencing of FAD2 and FAE1 genes affects both oleic acid and erucic acid contents in Brassica napus seeds[J]. Plant Cell Rep, 2010, 29(4): 317-325.
doi: 10.1007/s00299-010-0823-y pmid: 20130882 |
| [24] |
Hüsken A, Baumert A, Strack D, et al. Reduction of sinapate ester content in transgenic oilseed rape(Brassica napus)by dsRNAi-based suppression of BnSGT1 gene expression[J]. Mol Breed, 2005, 16(2): 127-138.
doi: 10.1007/s11032-005-6825-8 URL |
| [25] |
Padmanaban S, Lin XY, Perera I, et al. Differential expression of vacuolar H+-ATPase subunit c genes in tissues active in membrane trafficking and their roles in plant growth as revealed by RNAi[J]. Plant Physiol, 2004, 134(4): 1514-1526.
doi: 10.1104/pp.103.034025 pmid: 15051861 |
| [26] |
Subramanian S, Graham MY, Yu O, et al. RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and to enhanced susceptibility to Phytophthora sojae[J]. Plant Physiol, 2005, 137(4): 1345-1353.
doi: 10.1104/pp.104.057257 pmid: 15778457 |
| [27] | 陈艳, 柴友荣, 张迪, 等. 芸薹属TT19基因家族RNA干扰载体的构建[J]. 贵州农业科学, 2011, 39(3): 1-4. |
| Chen Y, Chai YR, Zhang D, et al. Construction of RNA interference vector of TT19 gene family in Brassica[J]. Guizhou Agric Sci, 2011, 39(3): 1-4. | |
| [28] |
He F, Ni N, Zeng ZY, et al. FAMSi:a synthetic biology approach to the fast assembly of multiplex siRNAs for silencing gene expression in mammalian cells[J]. Mol Ther Nucleic Acids, 2020, 22:885-899.
doi: 10.1016/j.omtn.2020.10.007 URL |
| [29] |
Wang X, Yuan CF, Huang B, et al. Developing a versatile shotgun cloning strategy for single-vector-based multiplex expression of short interfering RNAs(siRNAs)in mammalian cells[J]. ACS Synth Biol, 2019, 8(9): 2092-2105.
doi: 10.1021/acssynbio.9b00203 pmid: 31465214 |
| [30] | 陈任, 张虹, 张芮, 等. 用于植物多基因表达载体构建的质粒系统[J]. 分子植物育种, 2018, 16(4): 1138-1146. |
| Chen R, Zhang H, Zhang R, et al. A plasmid system for construction of plant multiple gene expression vectors[J]. Mol Plant Breed, 2018, 16(4): 1138-1146. | |
| [31] | 陈立志. 椰子FATB3、LPAAT和KASI多基因载体构建和协同作用分析[D]. 海口: 海南大学, 2018. |
| Chen LZ. Multi-gene vector assembly and co-expression analysis of FatB3, LPAAT and KASI from coconut[D]. Haikou: Hainan University, 2018. |
| [1] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [2] | LUO Hao-tian, WANG Long, WANG Yu-qian, WANG Yue, LI Jia-zhen, YANG Meng-ke, ZHANG Jie, DENG Xin, WANG Hong-yan. Genome-wide Identification and Expression Analysis of the RNAi-related Gene Families in Setaria viridis [J]. Biotechnology Bulletin, 2023, 39(1): 175-186. |
| [3] | SUN Wei, ZHANG Yan, WANG Yu-han, XU Hui, XU Xiao-rong, JU Zhi-gang. Cloning of Rd3GT1 in Rhododendron delavayi and Its Effect on Flower Color Formation of Petunia hybrida [J]. Biotechnology Bulletin, 2022, 38(9): 198-206. |
| [4] | YANG Jia-hui, SUN Yu-ping, LU Ya-ning, LIU huan, LU Cun-fu, CHEN Yu-zhen. Abiotic Stress Resistance of Escherichia coli Transformed with Arabidopsis thaliana AtTERT Gene [J]. Biotechnology Bulletin, 2022, 38(2): 1-9. |
| [5] | DANG Yuan, LI Wei, MIAO Xiang, XIU Yu, LIN Shan-zhi. Cloning of Oleosin Gene PsOLE4 from Prunus sibirica and Its Regulatory Function Analysis for Oil Accumulation [J]. Biotechnology Bulletin, 2022, 38(11): 151-161. |
| [6] | CUI Xiang-hua, TAO Nan, CHENG Bo-pu, ZHAO Yong-chang, CHEN Wei-min, LI Jing. Screening Promoters for Genetic Transformation of Cyclocybe aegerita [J]. Biotechnology Bulletin, 2021, 37(5): 259-266. |
| [7] | CHEN Ying, CHEN Xi, WANG Qian, WANG Xiao-li. Cloning,Expression and Biological Function Analysis of Universal Stress Protein in Festuca arundinacea [J]. Biotechnology Bulletin, 2021, 37(2): 32-39. |
| [8] | YANG Zhen-zhou, LIU Gang, XU Li. Reverse Transcription Digital PCR Detection Method for NAi-Based Transgenic Maize [J]. Biotechnology Bulletin, 2020, 36(5): 56-63. |
| [9] | YANG Wen-wen, NI Jia-yao, HU Rui-jie, WANG Hua-zhong. A Sequencing Strategy for Inverted Repeats in RNAi Vectors [J]. Biotechnology Bulletin, 2020, 36(5): 205-210. |
| [10] | XU Xiang, DONG Wei-peng, ZHANG Shao-hua, FENG Chen-yi, LIU Tian-fu, YAN Jiong. Construction of Fsp27 Gene Silencing Vector and Its Effect on Cell Lipolysis [J]. Biotechnology Bulletin, 2020, 36(1): 88-94. |
| [11] | LI Chong-hui, YIN Jun-mei. Genetic Engineering Progress and Breeding Tactics on Blue Flowers [J]. Biotechnology Bulletin, 2019, 35(11): 160-168. |
| [12] | ZHANG Ting, YANG Xuan-xuan, LEI Qin, LIU Juan-juan, LI Ji-gang. Effects of Phoxim,Methomyl and Feeding dsRNA on the Expression of ace Genes in Helicoverpa armigera [J]. Biotechnology Bulletin, 2018, 34(7): 160-165. |
| [13] | CHEN Jing, ZHANG Dao-wei, QIAN Zheng-min. Structure and Characteristics Analysis of Chitin Synthase 1 from Sogota furcifera [J]. Biotechnology Bulletin, 2018, 34(1): 195-201. |
| [14] | GUO Hong-yan, GAO Han, WU Qi, SUN Xiao-jie, LIU Xiu-cai, ZHAO Li-qun. Construction of SGK3 Gene Lentiviral RNA Interference Vector and Effects on Cell proliferation and Apoptosis of Breast Cancer Cell Line MB-474 [J]. Biotechnology Bulletin, 2018, 34(1): 247-252. |
| [15] | FENG Xiao-yan ZHANG Shu-zhen. Research Advances on RNAi Mechanism and Its Application [J]. Biotechnology Bulletin, 2017, 33(5): 1-8. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||