








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (1): 157-165.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0560
Previous Articles Next Articles
LIN Rong1( ), ZHENG Yue-ping2(
), ZHENG Yue-ping2( ), XU Xue-zhen2, LI Dan-dan2, ZHENG Zhi-fu1,2
), XU Xue-zhen2, LI Dan-dan2, ZHENG Zhi-fu1,2
Received:2022-05-09
Online:2023-01-26
Published:2023-02-02
Contact:
ZHENG Yue-ping
E-mail:lynerlin@163.com;zyp860819@126.com
LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana[J]. Biotechnology Bulletin, 2023, 39(1): 157-165.
| 靶基因 Target gene | sgRNA序列 sgRNA sequence(5'-3') | 位置 Location |
|---|---|---|
| ACOL8 | TCTTTCGAGGAGACTATGACAGG | 外显子(1) |
| ACOL8 | GCACGTCGAGCGGGATCCCGTGG | 外显子(1) |
Table 1 Target sequences for ACOL8 gene editing
| 靶基因 Target gene | sgRNA序列 sgRNA sequence(5'-3') | 位置 Location |
|---|---|---|
| ACOL8 | TCTTTCGAGGAGACTATGACAGG | 外显子(1) |
| ACOL8 | GCACGTCGAGCGGGATCCCGTGG | 外显子(1) |
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 用途 Utility |
|---|---|---|
| ZYP209-BsF | ATATATggtctcGATTGCTTTCGA- GGAGACTATGACGTT | 构建sgRNA表达盒 |
| ZYP210-F0 | TGCTTTCGAGGAGACTATGAC- GTTTTAGAGCTAGAAATAGC | 构建sgRNA表达盒 |
| ZYP211-R0 | AACCGGGATCCCGCTCGACGT- GCAATCTCTTAGTCGACTCTAC | 构建sgRNA表达盒 |
| ZYP212-BsR | ATTATTggtctcGAAACCGGGAT- CCCGCTCGACGTGC | 构建sgRNA表达盒 |
| ZYP237-FP | ATTCCTTTGATGCCGTGATAGT | 鉴定筛选,测序 |
| ZYP238-RP | TAGCTGGAACCTCTTTGATTCC | 鉴定筛选 |
| ZYP239-FP | ATCGATCTGAACGGAGGAGTAG | 鉴定筛选 |
| ZYP240-RP | TTTCGAGTGTGATCACGAGAGT | 鉴定筛选,测序 |
Table 2 Primer sequences
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 用途 Utility |
|---|---|---|
| ZYP209-BsF | ATATATggtctcGATTGCTTTCGA- GGAGACTATGACGTT | 构建sgRNA表达盒 |
| ZYP210-F0 | TGCTTTCGAGGAGACTATGAC- GTTTTAGAGCTAGAAATAGC | 构建sgRNA表达盒 |
| ZYP211-R0 | AACCGGGATCCCGCTCGACGT- GCAATCTCTTAGTCGACTCTAC | 构建sgRNA表达盒 |
| ZYP212-BsR | ATTATTggtctcGAAACCGGGAT- CCCGCTCGACGTGC | 构建sgRNA表达盒 |
| ZYP237-FP | ATTCCTTTGATGCCGTGATAGT | 鉴定筛选,测序 |
| ZYP238-RP | TAGCTGGAACCTCTTTGATTCC | 鉴定筛选 |
| ZYP239-FP | ATCGATCTGAACGGAGGAGTAG | 鉴定筛选 |
| ZYP240-RP | TTTCGAGTGTGATCACGAGAGT | 鉴定筛选,测序 |
| 引物名称 Primer name | 基因 Gene | 用途 Purpose | 引物序列 Primer sequence(5'-3') |
|---|---|---|---|
| ZZ200 qPCR-FP | ACTIN | 内参基因 | GTCGTACAACCGGTATTGTGCT |
| ZZ201 qPCR-RP | TGTCTCTTACAATTTCCCGCTCT | ||
| ZYP197 qPCR-FP | ACOL8 | 目标基因 | GGGGCTCTTGTCGTTAACCT |
| ZYP198 qPCR-RP | TCCATATACTCGATGGCTCTCC |
Table 3 Sequences of the primers used in real-time fluore-scent quantitative PCR
| 引物名称 Primer name | 基因 Gene | 用途 Purpose | 引物序列 Primer sequence(5'-3') |
|---|---|---|---|
| ZZ200 qPCR-FP | ACTIN | 内参基因 | GTCGTACAACCGGTATTGTGCT |
| ZZ201 qPCR-RP | TGTCTCTTACAATTTCCCGCTCT | ||
| ZYP197 qPCR-FP | ACOL8 | 目标基因 | GGGGCTCTTGTCGTTAACCT |
| ZYP198 qPCR-RP | TCCATATACTCGATGGCTCTCC |
| 基因 Gene | 基因ID Gene ID | 编码蛋白 Encoded protein | 氨基酸 Amino acid/aa |
|---|---|---|---|
| ACO1 | AT2G19590 | ACO1 | 310 |
| ACO2 | AT1G62380 | ACO2 | 320 |
| ACO3 | AT1G12010 | ACO3 | 320 |
| ACO4 | AT1G05010 | ACO4 | 323 |
| ACO5 | AT1G77330 | ACO5 | 307 |
| ACOL1 | AT1G06620 | ACO-like homolog 1 | 365 |
| ACOL2 | AT1G06640 | ACO-like homolog 2 | 369 |
| ACOL3 | AT1G06650 | ACO-like homolog 3 | 369 |
| ACOL4 | AT1G03400 | ACO-like homolog 4 | 351 |
| ACOL5 | AT1G03410 | ACO-like homolog 5 | 398 |
| ACOL6 | AT1G04350 | ACO-like homolog 6 | 360 |
| ACOL7 | AT1G04380 | ACO-like homolog 7 | 345 |
| ACOL8 | AT3G61400 | ACO-like homolog 8 | 370 |
| ACOL9 | AT5G43440 | ACO-like homolog 9 | 365 |
| ACOL10 | AT5G43450 | ACO-like homolog 10 | 362 |
| ACOL11 | AT5G59530 | ACO-like homolog 11 | 364 |
| ACOL12 | AT5G59540 | ACO-like homolog 12 | 366 |
Table 4 Characteristics of genes encoding Arabidopsis ACC oxidases and ACO-like homologs
| 基因 Gene | 基因ID Gene ID | 编码蛋白 Encoded protein | 氨基酸 Amino acid/aa |
|---|---|---|---|
| ACO1 | AT2G19590 | ACO1 | 310 |
| ACO2 | AT1G62380 | ACO2 | 320 |
| ACO3 | AT1G12010 | ACO3 | 320 |
| ACO4 | AT1G05010 | ACO4 | 323 |
| ACO5 | AT1G77330 | ACO5 | 307 |
| ACOL1 | AT1G06620 | ACO-like homolog 1 | 365 |
| ACOL2 | AT1G06640 | ACO-like homolog 2 | 369 |
| ACOL3 | AT1G06650 | ACO-like homolog 3 | 369 |
| ACOL4 | AT1G03400 | ACO-like homolog 4 | 351 |
| ACOL5 | AT1G03410 | ACO-like homolog 5 | 398 |
| ACOL6 | AT1G04350 | ACO-like homolog 6 | 360 |
| ACOL7 | AT1G04380 | ACO-like homolog 7 | 345 |
| ACOL8 | AT3G61400 | ACO-like homolog 8 | 370 |
| ACOL9 | AT5G43440 | ACO-like homolog 9 | 365 |
| ACOL10 | AT5G43450 | ACO-like homolog 10 | 362 |
| ACOL11 | AT5G59530 | ACO-like homolog 11 | 364 |
| ACOL12 | AT5G59540 | ACO-like homolog 12 | 366 |
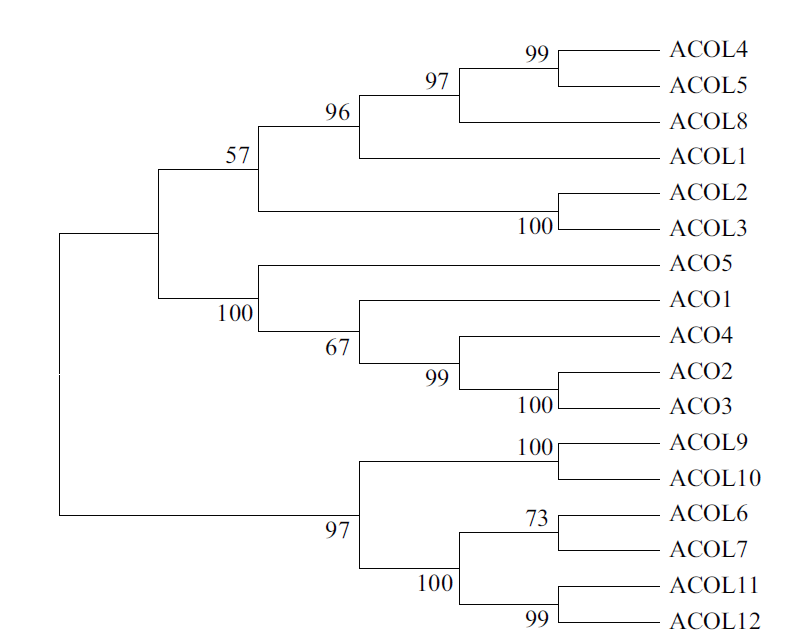
Fig. 2 A dendrogram analysis of members of the ACC oxidase family and ACO-like homologs The MEGA7 software groups the sequences according to the results of amino acid sequence alignment,and the proteins classified into the same group have a relatively close relationship

Fig. 3 Amino acid sequences alignment of the ACC oxidase family members and ACOL8 Amino acid sites with the same color indicate homology,black=100%,red>75%,green>50%;sgRNA refers to the amino acid sequence corresponding to the gene editing site
| 株系Line | 突变体编号Mutant code | 突变类型Mutation type | 突变位点Mutation site | 突变特点Mutation characteristics |
|---|---|---|---|---|
| acol8-1 | A10/WT-14-5-38 | 插入 | 308-309:5 bp插入CTGGA | 移码突变 |
| acol8-2 | A10/WT-230-17 | 缺失 | 66-308:243 bp缺失 | 大片段缺失 |
| acol8-3 | A10/WT-254-19 | 插入 | 65-66:1 bp插入T 308-309:1 bp插入T | 提前终止 |
Table 5 Mutation sites in the three acol8 mutants
| 株系Line | 突变体编号Mutant code | 突变类型Mutation type | 突变位点Mutation site | 突变特点Mutation characteristics |
|---|---|---|---|---|
| acol8-1 | A10/WT-14-5-38 | 插入 | 308-309:5 bp插入CTGGA | 移码突变 |
| acol8-2 | A10/WT-230-17 | 缺失 | 66-308:243 bp缺失 | 大片段缺失 |
| acol8-3 | A10/WT-254-19 | 插入 | 65-66:1 bp插入T 308-309:1 bp插入T | 提前终止 |
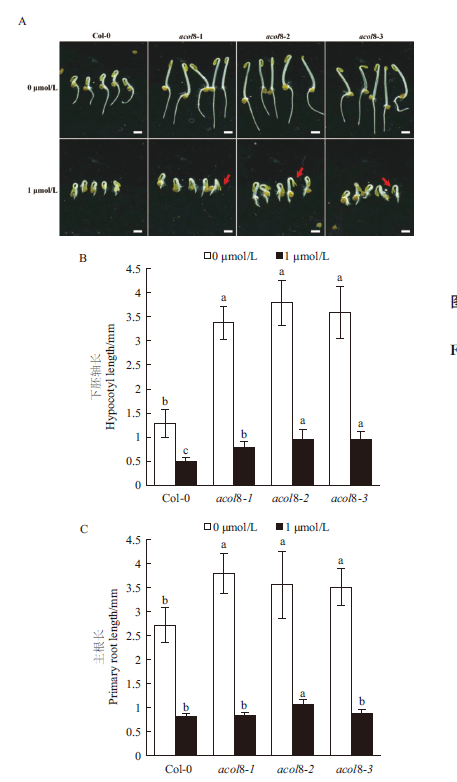
Fig. 5 Comparison of the “triple responses” of etiolated seedlings of wild type Arabidopsis and acol8 mutant A:The phenotype of etiolated seedlings of acol8 single mutant and wild type Arabidopsis treated with different concentrations of ACC,Bar = 2 mm. B:Length of hypocotyl of etiolated seedlings of acol8 single mutants and wild-type Arabidopsis treated with different concentrations of ACC. C:Length of primary root of etiolated seedlings of acol8 single mutants and wild type Arabidopsis. Different letters indicate significant differences at the 0.05 level,the same below
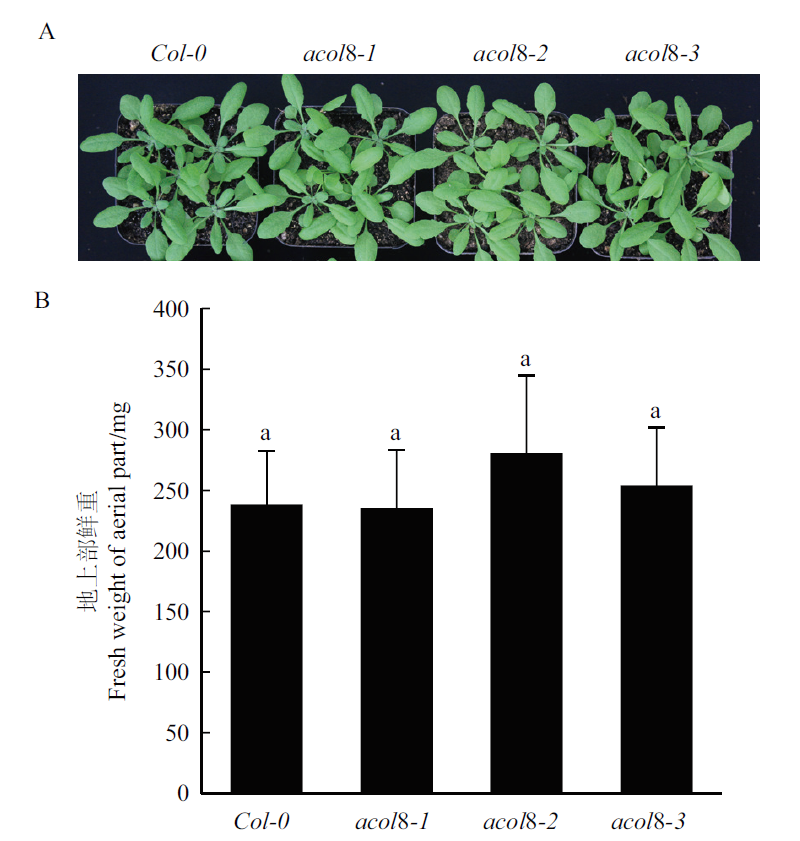
Fig. 7 Comparison of the phenotypes of wild type Arabid-opsis and three acol8 mutants A:The aboveground phenotype of wild type Arabidopsis and acol8 single mutant,Bar=1 cm. B:The fresh weight of aboveground biomass of wild type Arabidopsis and acol8 single mutants
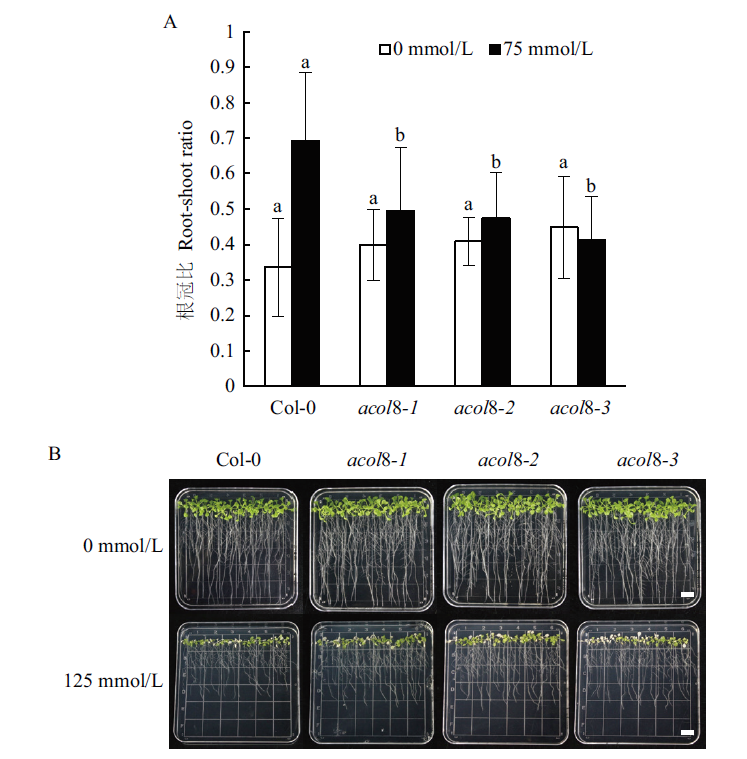
Fig. 8 Comparison of salt tolerances of wild type Arabido-psis and acol8 mutants Comparison of the root-shoot ratio (A)and the phenotype (B)of wild type Arabi-dopsis with that of acol8 mutant under different concentrations of NaCl,Bar=1 cm
| [1] |
Ahammed GJ, Gantait S, Mitra M, et al. Role of ethylene crosstalk in seed germination and early seedling development:a review[J]. Plant Physiol Biochem, 2020, 151:124-131.
doi: 10.1016/j.plaphy.2020.03.016 URL |
| [2] |
Nascimento VL, Pereira AM, Pereira AS, et al. Physiological and metabolic bases of increased growth in the tomato ethylene-insensitive mutant Never ripe:extending ethylene signaling functions[J]. Plant Cell Rep, 2021, 40(8): 1377-1393.
doi: 10.1007/s00299-020-02623-y pmid: 33074436 |
| [3] |
Tripathi SK, Tuteja N. Integrated signaling in flower senescence[J]. Plant Signal Behav, 2007, 2(6): 437-445.
doi: 10.4161/psb.2.6.4991 URL |
| [4] |
Dubois M, van den Broeck L, Inzé D. The pivotal role of ethylene in plant growth[J]. Trends Plant Sci, 2018, 23(4): 311-323.
doi: S1360-1385(18)30015-3 pmid: 29428350 |
| [5] | Tao JJ, Chen HW, Ma B, et al. The role of ethylene in plants under Sal Inity stress[J]. Front Plant Sci, 2015, 6:1059. |
| [6] |
Kazan K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance[J]. Trends Plant Sci, 2015, 20(4): 219-229.
doi: 10.1016/j.tplants.2015.02.001 pmid: 25731753 |
| [7] |
Yu YB, Adams DO, Yang SF. 1-Aminocyclopropanecarboxylate synthase, a key enzyme in ethylene biosynthesis[J]. Arch Biochem Biophys, 1979, 198(1): 280-286.
pmid: 507845 |
| [8] |
Adams DO, Yang SF. Ethylene biosynthesis:identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene[J]. Proc Natl Acad Sci USA, 1979, 76(1): 170-174.
doi: 10.1073/pnas.76.1.170 pmid: 16592605 |
| [9] | Park CH, Roh J, Youn JH, et al. Arabidopsis ACC oxidase 1 coordinated by multiple signals mediates ethylene biosynthesis and is involved in root development[J]. Mol Cells, 2018, 41(10): 923-932. |
| [10] |
Ning Q, Jian YN, Du YF, et al. An ethylene biosynthesis enzyme controls quantitative variation in maize ear length and kernel yield[J]. Nat Commun, 2021, 12(1): 5832.
doi: 10.1038/s41467-021-26123-z pmid: 34611160 |
| [11] |
Chersicola M, Kladnik A, Žnidarič MT, et al. 1-aminocyclopropane-1-carboxylate oxidase induction in tomato flower pedicel phloem and abscission related processes are differentially sensitive to ethylene[J]. Front Plant Sci, 2017, 8:464.
doi: 10.3389/fpls.2017.00464 pmid: 28408916 |
| [12] |
Penarrubia L, Aguilar M, Margossian L, et al. An antisense gene stimulates ethylene hormone production during tomato fruit ripening[J]. Plant Cell, 1992, 4(6): 681-687.
doi: 10.2307/3869526 URL |
| [13] |
Lincoln JE, Fischer RL. Diverse mechanisms for the regulation of ethylene-inducible gene expression[J]. Mol Gen Genet, 1988, 212(1): 71-75.
doi: 10.1007/BF00322446 URL |
| [14] |
Wang ZP, Xing HL, Dong L, et al. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation[J]. Genome Biol, 2015, 16(1): 144.
doi: 10.1186/s13059-015-0715-0 URL |
| [15] | 朱丽颖, 郑月萍, 徐雪珍, 等. 一种准确、简便测定CRISPR/Cas9基因编辑效率的方法[J]. 江苏农业学报, 2020, 36(2): 299-305. |
| Zhu LY, Zheng YP, Xu XZ, et al. A convenient and accurate method for determining the efficiency of CRISPR/Cas9-based gene editing[J]. Jiangsu J Agric Sci, 2020, 36(2): 299-305. | |
| [16] | Giovannoni JJ. Genetic regulation of fruit development and ripening[J]. Plant Cell, 2004, 16(Suppl):S170-S180. |
| [17] |
Yin XR, Allan AC, Chen KS, et al. Kiwifruit EIL and ERF genes involved in regulating fruit ripening[J]. Plant Physiol, 2010, 153(3): 1280-1292.
doi: 10.1104/pp.110.157081 URL |
| [18] |
Riyazuddin R, Verma R, Singh K, et al. Ethylene:a master regulator of Sal Inity stress tolerance in plants[J]. Biomolecules, 2020, 10(6): 959.
doi: 10.3390/biom10060959 URL |
| [19] |
Jiang CF, Belfield EJ, Cao Y, et al. An Arabidopsis soil-Sal Inity-tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis[J]. Plant Cell, 2013, 25(9): 3535-3552.
doi: 10.1105/tpc.113.115659 URL |
| [20] |
Lockhart J. Salt of the earth:ethylene promotes salt tolerance by enhancing Na/K homeostasis[J]. Plant Cell, 2013, 25(9): 3150.
doi: 10.1105/tpc.113.250911 URL |
| [1] | HAN Zhi-yang, JIA Zi-miao, LIANG Qiu-ju, WANG Ke, TANG Hua-li, YE Xing-guo, ZHANG Shuang-xi. Salt Tolerance at Seedling Stage and Analysis of Selenium and Folic Acid Content in Seeds in Two Sets of Wheat-Dasypyrum villosum Chromosom Additional Lines [J]. Biotechnology Bulletin, 2023, 39(8): 185-193. |
| [2] | LI Yu, LI Su-zhen, CHEN Ru-mei, LU Hai-qiang. Advances in the Regulation of Iron Homeostasis by bHLH Transcription Factors in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 26-36. |
| [3] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [4] | CHEN Yi-bo, YANG Wan-ming, YUE Ai-qin, WANG Li-xiang, DU Wei-jun, WANG Min. Construction of Soybean Genetic Map Based on SLAF Markers and QTL Mapping Analysis of Salt Tolerance at Seedling Stage [J]. Biotechnology Bulletin, 2023, 39(2): 70-79. |
| [5] | LIU Jia-xin, ZHANG Hui-long, ZOU Rong-song, YANG Xiu-yan, ZHU Jian-feng, ZHANG Hua-xin. Research Progress in Na+ Antiport and Physiological Growth Mechanisms of Differernt Halophytes Adapted to Salt Stress [J]. Biotechnology Bulletin, 2023, 39(1): 59-72. |
| [6] | CHEN Hong-yan, LI Xiao-er, LI Zhong-guang. Sugar Signaling and Its Role in Plant Response to Environmental Stress [J]. Biotechnology Bulletin, 2022, 38(7): 80-89. |
| [7] | YIPARE·Paerhati , ZULIHUMAER·Rouzi , TIAN Yong-zhi, ZHU Yan-lei, LI Yuan-ting, MA Xiao-lin. Research Progress in Diversity of Endophytes Microbial Communities Isolated from Desert Plants and Their Strengthening Effects on Drought and Salt Tolerance in Crops [J]. Biotechnology Bulletin, 2022, 38(12): 88-99. |
| [8] | LI Cai-xia, LAN Hai-yan. Research Progress in the Stress Tolerance Mechanisms of Desert Plant Tamarix spp. [J]. Biotechnology Bulletin, 2021, 37(5): 128-140. |
| [9] | YANG Hua-jie, ZHOU Yu-ping, FAN Tian, LV Tian-xiao, XIE Chu-ping, TIAN Chang-en. Screening and Identification of IQM4-Interacting Proteins in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2021, 37(11): 190-196. |
| [10] | FANG Dan-dan, ZHANG Ting, WEN Xiao-peng. Overexpression of Pinus massoniana PmPT3 Gene in Arabidopsis thaliana Increasing Low Phosphorus Tolerance [J]. Biotechnology Bulletin, 2021, 37(10): 1-8. |
| [11] | HU Yu-jie, ZHU Xiu-ling, DING Yan-qin, DU Bing-hai, WANG Cheng-qiang. Research Progress on Salt Tolerance and Growth-promoting Mechanism of Bacillus [J]. Biotechnology Bulletin, 2020, 36(9): 64-74. |
| [12] | ZHANG Xiao-jia, LU Ya-jun, ZHANG Wen-jin, ZHANG Yu, CUI Gao-chang, LANG Duo-yong, ZHANG Xin-hui. Preparation of Drought-resistant and Salt-tolerant Bacteria and Its Effect on Germination of Licorice Seeds [J]. Biotechnology Bulletin, 2020, 36(9): 180-193. |
| [13] | SHI Jing-jing, GUO Yi-ping, YU Ying, ZHOU Mei-qi, WANG Chao. Analysis of Salt Tolerance of Transgenic BpLTP4 Tobacco [J]. Biotechnology Bulletin, 2020, 36(12): 34-41. |
| [14] | REN Lei, LIU Bin, LIN Zhong, ZHEN Zhen, LIU Yue-lian, HU Han-qiao, YAN Yan-chun. Isolation of a p-Nitrophenol-Degrading Bacterium and Investigation of Its Degrading Mechanism [J]. Biotechnology Bulletin, 2019, 35(9): 184-193. |
| [15] | ZHANG Zhao-yang, PANG Jun-ling, HAN Mei, LENG Peng-fei, ZHAO Jun. Characterization of the Salt Tolerance of Transgenic Maize Line Expressing ABP9 [J]. Biotechnology Bulletin, 2019, 35(5): 48-57. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||