








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (3): 152-162.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0974
Previous Articles Next Articles
XIE Yang1,2( ), XING Yu-meng1,2, ZHOU Guo-yan1,2, LIU Mei-yan1,2, YIN Shan-shan1,2, YAN Li-ying1,2
), XING Yu-meng1,2, ZHOU Guo-yan1,2, LIU Mei-yan1,2, YIN Shan-shan1,2, YAN Li-ying1,2
Received:2022-08-09
Online:2023-03-26
Published:2023-04-10
XIE Yang, XING Yu-meng, ZHOU Guo-yan, LIU Mei-yan, YIN Shan-shan, YAN Li-ying. Transcriptome Analysis of Diploid and Autotetraploid in Cucumber Fruit[J]. Biotechnology Bulletin, 2023, 39(3): 152-162.
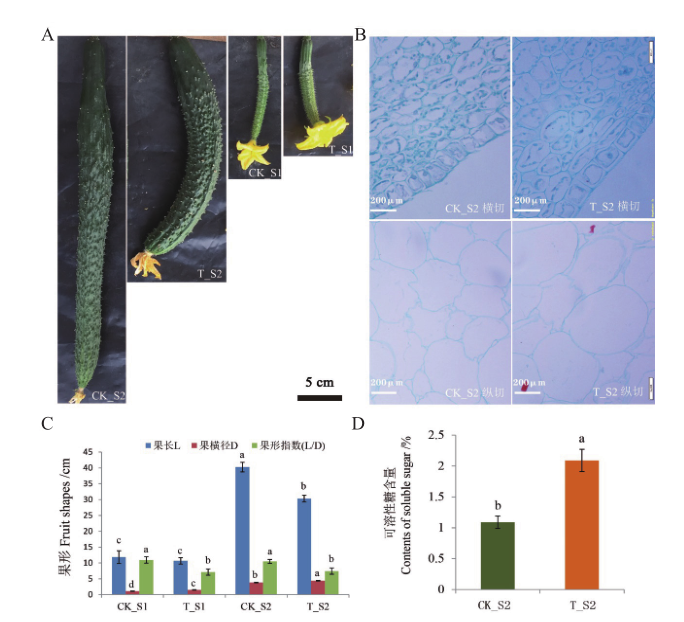
Fig. 1 Analysis of diploid and autotetraploid fruit shapes as well as their soluble sugar contents in cucumber A: Morphological observation of cucumber fruits at different developmental stages of diploid and tetraploid. B: Fruit cytology of cucumber diploid and tetraploid at different developmental stages. C: Statistics of fruit shape index of cucumber diploid and tetraploid at different developmental stages. D: Analysis of soluble sugar content in diploid and tetraploid fruits of cucumber. Different lower letters indicate significant difference at 0.05 level
| 样品名称 Sample name | Raw reads/Mb | Clean reads/Mb | Unique map/Mb | Q30/% | GC/% |
|---|---|---|---|---|---|
| CK_S1_1 | 45.32 | 43.78 | 40.87 | 92.94 | 40.88 |
| CK_S1_2 | 48.02 | 45.29 | 43.08 | 94.72 | 40.85 |
| CK_S2_1 | 44.23 | 43.35 | 40.80 | 93.02 | 42.14 |
| CK_S2_2 | 44.89 | 43.22 | 40.68 | 92.63 | 42.28 |
| T_S1_1 | 46.21 | 43.69 | 40.35 | 92.56 | 40.82 |
| T_S1_2 | 47.89 | 45.85 | 42.61 | 92.98 | 41.09 |
| T_S2_1 | 45.83 | 44.53 | 42.08 | 92.9 | 42.43 |
| T_S2_2 | 47.69 | 45.88 | 43.46 | 93.49 | 42.43 |
Table 1 Quality analysis of transcriptome sequencing data
| 样品名称 Sample name | Raw reads/Mb | Clean reads/Mb | Unique map/Mb | Q30/% | GC/% |
|---|---|---|---|---|---|
| CK_S1_1 | 45.32 | 43.78 | 40.87 | 92.94 | 40.88 |
| CK_S1_2 | 48.02 | 45.29 | 43.08 | 94.72 | 40.85 |
| CK_S2_1 | 44.23 | 43.35 | 40.80 | 93.02 | 42.14 |
| CK_S2_2 | 44.89 | 43.22 | 40.68 | 92.63 | 42.28 |
| T_S1_1 | 46.21 | 43.69 | 40.35 | 92.56 | 40.82 |
| T_S1_2 | 47.89 | 45.85 | 42.61 | 92.98 | 41.09 |
| T_S2_1 | 45.83 | 44.53 | 42.08 | 92.9 | 42.43 |
| T_S2_2 | 47.69 | 45.88 | 43.46 | 93.49 | 42.43 |
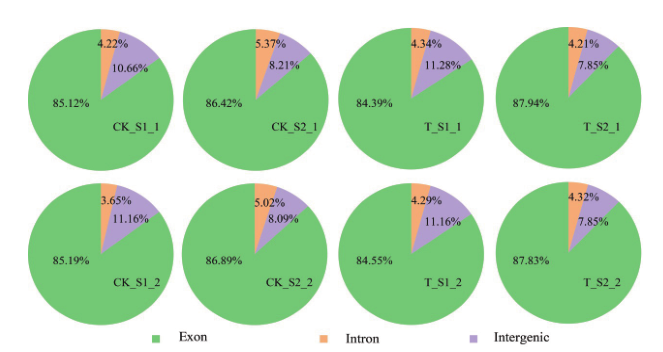
Fig. 2 Percents of clean reads mapped in genome regions Distribution of clean reads sequenced in different developmental stages of cucumber diploid and tetraploid fruits in genomic regions
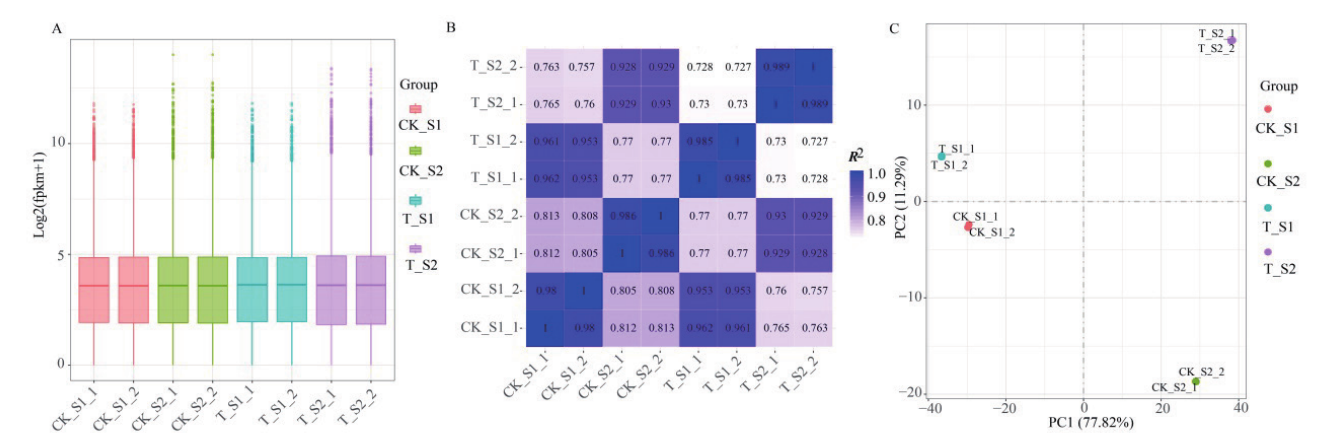
Fig. 3 Gene expression distribution and correlation analysis of samples A: Box diagram of gene expression distribution in cucumber diploid and tetraploid fruits at different developmental stages. B: Pearson correlation analysis of cucumber diploid and tetraploid fruit samples at different developmental stages. C: PCA analysis of gene expression levels in diploid and tetraploid cucumber fruits at different developmental stages
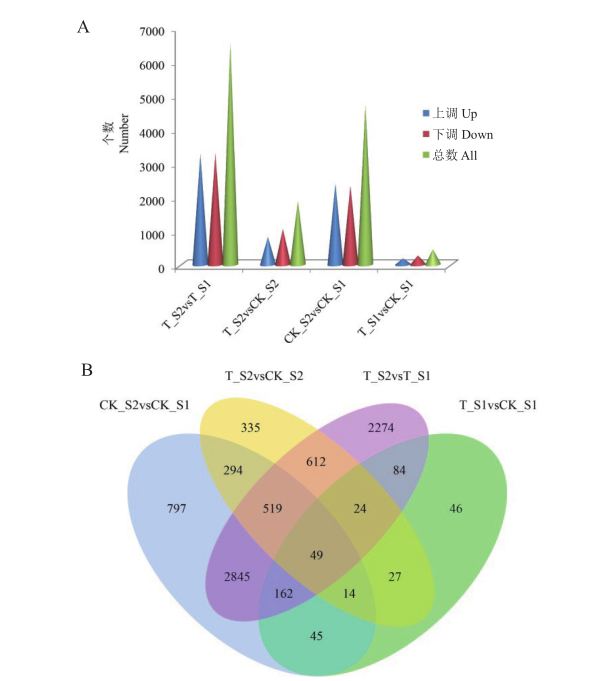
Fig. 4 Analysis of DEGs statistical number and Venn diagram A: Bar chart of DEGs statistics of different comparison groups in different developmental stages of cucumber diploid and tetraploid fruits. B: Venn diagram of DEGs in different comparison groups at different developmental stages of cucumber diploid and tetraploid fruits
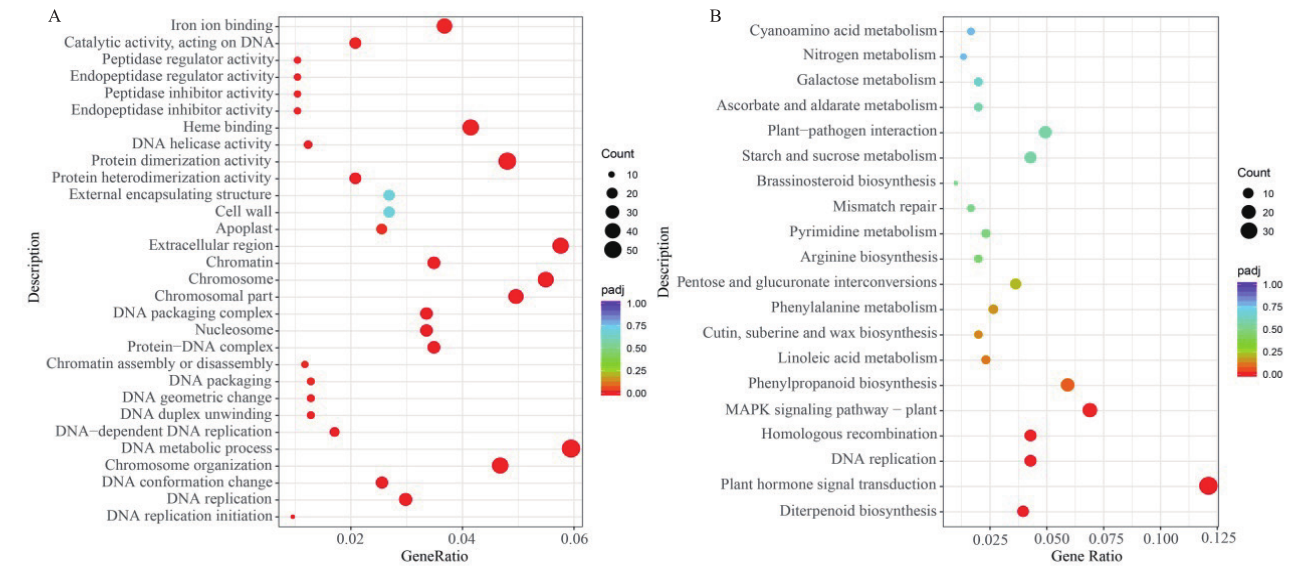
Fig. 5 Rich distribution point diagram for GO and KEGG A: Map of GO enrichment of DEGs in T_S2vsCK_S2 of cucumber fruit. B: Map of KEGG enrichment of DEGs in T_S2vsCK_S2 of cucumber fruit
| 基因名称Gene name | 基因 ID Gene ID | log2FC | 基因注释Gene annotation |
|---|---|---|---|
| AUX1 | Csa_3G731880 | 0.511028525 | Auxin influx carrier(AUX1 LAX family) |
| B-ARR | Csa_2G350290 | 1.199233905 | Uncharacterized protein LOC101203501 isoform X1 |
| DELLA | Csa_5G569350 | 0.641993478 | DELLA protein |
| SIMKK | Csa_3G651720 | 0.521551332 | Mitogen-activated protein kinase kinase 4/5 |
| EIN2 | Csa_6G445230 | 0.401357885 | Ethylene-insensitive protein 2 |
| EIN3 | Csa_6G051520 | 0.672607401 | Ethylene-insensitive protein 3 |
| BRI1 | Csa_6G486870 | 0.529640378 | Protein brassinosteroid insensitive 1 |
| BIN2 | Csa_6G157100 | 0.536467357 | Protein brassinosteroid insensitive 2 |
| BZR1/2 | Csa_2G361450 | 0.587660284 | Brassinosteroid resistant 1/2 |
| TCH4 | Csa_1G422470 | 1.185601828 | Xyloglucan:xyloglucosyl transferase TCH4 |
| TGA | Csa_6G152350 | 1.955263915 | Transcription factor TGA |
| GH3 | Csa_6G493310 | -0.840958686 | Auxin responsive GH3 gene family |
| GID1 | Csa_7G391240 | -1.403103089 | Gibberellin receptor GID1 |
| PP2C | Csa_1G574880 | -1.528004155 | Protein phosphatase 2C |
| ETR | Csa_2G070880 | -0.799320186 | Ethylene receptor |
| BAK1 | Csa_7G060160 | -0.875573337 | Brassinosteroid insensitive 1-associated receptor kinase 1 |
| BKI1 | Csa_2G359970 | -1.610933985 | BRI1 kinase inhibitor 1 |
| CYCD3 | Csa_6G491650 | -1.00709524 | Cyclin D3, plant |
| JAZ | Csa_1G042920 | -0.913564833 | Jasmonate ZIM domain-containing protein |
| AGLU1 | Csa_1G716250 | 0.650665663 | Alpha-glucosidase |
| SPS | Csa_2G401440 | 0.977042232 | Sucrose-phosphate synthase |
| SUSY | Csa_5G322500 | 2.088838655 | Sucrose synthase |
| FRK | Csa_3G345390 | -0.650140834 | Fuctokinase |
| AGPase | Csa_3G149890 | -1.393581098 | Glucose-1-phosphate adenylyltransferase |
| SS | Csa_6G497160 | -0.899393235 | Starch synthase |
| GTase | Csa_5G606600 | -0.684711983 | 4-alpha-glucanotransferase |
| GBE1 | Csa_7G213190 | -1.304743764 | 1,4-alpha-glucan branching enzyme |
| BMY | Csa_3G017020 | -3.271484788 | Beta-amylase |
| TRE | Csa_5G149880 | -1.411978481 | Alpha,alpha-trehalase |
Table 2 Key candidate genes in KEGG enrichment pathway
| 基因名称Gene name | 基因 ID Gene ID | log2FC | 基因注释Gene annotation |
|---|---|---|---|
| AUX1 | Csa_3G731880 | 0.511028525 | Auxin influx carrier(AUX1 LAX family) |
| B-ARR | Csa_2G350290 | 1.199233905 | Uncharacterized protein LOC101203501 isoform X1 |
| DELLA | Csa_5G569350 | 0.641993478 | DELLA protein |
| SIMKK | Csa_3G651720 | 0.521551332 | Mitogen-activated protein kinase kinase 4/5 |
| EIN2 | Csa_6G445230 | 0.401357885 | Ethylene-insensitive protein 2 |
| EIN3 | Csa_6G051520 | 0.672607401 | Ethylene-insensitive protein 3 |
| BRI1 | Csa_6G486870 | 0.529640378 | Protein brassinosteroid insensitive 1 |
| BIN2 | Csa_6G157100 | 0.536467357 | Protein brassinosteroid insensitive 2 |
| BZR1/2 | Csa_2G361450 | 0.587660284 | Brassinosteroid resistant 1/2 |
| TCH4 | Csa_1G422470 | 1.185601828 | Xyloglucan:xyloglucosyl transferase TCH4 |
| TGA | Csa_6G152350 | 1.955263915 | Transcription factor TGA |
| GH3 | Csa_6G493310 | -0.840958686 | Auxin responsive GH3 gene family |
| GID1 | Csa_7G391240 | -1.403103089 | Gibberellin receptor GID1 |
| PP2C | Csa_1G574880 | -1.528004155 | Protein phosphatase 2C |
| ETR | Csa_2G070880 | -0.799320186 | Ethylene receptor |
| BAK1 | Csa_7G060160 | -0.875573337 | Brassinosteroid insensitive 1-associated receptor kinase 1 |
| BKI1 | Csa_2G359970 | -1.610933985 | BRI1 kinase inhibitor 1 |
| CYCD3 | Csa_6G491650 | -1.00709524 | Cyclin D3, plant |
| JAZ | Csa_1G042920 | -0.913564833 | Jasmonate ZIM domain-containing protein |
| AGLU1 | Csa_1G716250 | 0.650665663 | Alpha-glucosidase |
| SPS | Csa_2G401440 | 0.977042232 | Sucrose-phosphate synthase |
| SUSY | Csa_5G322500 | 2.088838655 | Sucrose synthase |
| FRK | Csa_3G345390 | -0.650140834 | Fuctokinase |
| AGPase | Csa_3G149890 | -1.393581098 | Glucose-1-phosphate adenylyltransferase |
| SS | Csa_6G497160 | -0.899393235 | Starch synthase |
| GTase | Csa_5G606600 | -0.684711983 | 4-alpha-glucanotransferase |
| GBE1 | Csa_7G213190 | -1.304743764 | 1,4-alpha-glucan branching enzyme |
| BMY | Csa_3G017020 | -3.271484788 | Beta-amylase |
| TRE | Csa_5G149880 | -1.411978481 | Alpha,alpha-trehalase |
| 基因名称Gene name | 注释Annotation | 引物序列Primer sequence(5'-3') |
|---|---|---|
| SUSY | 蔗糖合酶 Sucrose synthase | F:TGAGAACGATGAGCATATAG |
| R:TCCAACCACAACAAGATT | ||
| K-box | 转录因子K-box Transcription factor K-box | F:CAAGAACAGGAAGAGGAA |
| R:GAGGAGAAGACGATAAGAG | ||
| WUS | WUSCHEL相关homeobox WUSCHEL-related homeobox | F:GTGAGCCATTGATGACAT |
| R:GGAACCAGTTATAGACATTAGA | ||
| SUT | 蔗糖转运蛋白 Sucrose transporter | F:CTGAGACGGTTACTAAGAG |
| R:TTGAATATAAGGAGTGAGAAGA | ||
| GLU8 | 葡萄糖转运蛋白8 Glucose transporter protein 8 | F:CCTCACAACACTCAATCA |
| R:CACAGACCAACAATCAGA | ||
| DNAH | DNA解旋酶 DNA helicase | F:GCTCAAGTCGTTACCATT |
| R:CATCAACTGAACTCACCTT | ||
| ARF4 | 激素响应因子4 Auxin response factor 4 | F:GTCTACCATACTCGTGTT |
| R:CTAATGCCAGTGATTGTG | ||
| GIR | 赤霉素调控蛋白 Gibberellin-regulated protein | F:TCCTCTCTTCTCGTTCTT |
| R:GCTCCTCCACAATCAATA | ||
| LRK | 蛋白激酶家族蛋白 Protein kinase family protein | F:CGGAGATGACTGTTGTAA |
| R:TCCAATCAGCAATAATAACG | ||
| CsActin | 内参基因 Actin gene | F:ATTGTTCTCAGTGGTGGTTCTAC |
| R:CCTTTGAGATCCACATCTGCT |
Table 3 Information of RT-qPCR primer sequences
| 基因名称Gene name | 注释Annotation | 引物序列Primer sequence(5'-3') |
|---|---|---|
| SUSY | 蔗糖合酶 Sucrose synthase | F:TGAGAACGATGAGCATATAG |
| R:TCCAACCACAACAAGATT | ||
| K-box | 转录因子K-box Transcription factor K-box | F:CAAGAACAGGAAGAGGAA |
| R:GAGGAGAAGACGATAAGAG | ||
| WUS | WUSCHEL相关homeobox WUSCHEL-related homeobox | F:GTGAGCCATTGATGACAT |
| R:GGAACCAGTTATAGACATTAGA | ||
| SUT | 蔗糖转运蛋白 Sucrose transporter | F:CTGAGACGGTTACTAAGAG |
| R:TTGAATATAAGGAGTGAGAAGA | ||
| GLU8 | 葡萄糖转运蛋白8 Glucose transporter protein 8 | F:CCTCACAACACTCAATCA |
| R:CACAGACCAACAATCAGA | ||
| DNAH | DNA解旋酶 DNA helicase | F:GCTCAAGTCGTTACCATT |
| R:CATCAACTGAACTCACCTT | ||
| ARF4 | 激素响应因子4 Auxin response factor 4 | F:GTCTACCATACTCGTGTT |
| R:CTAATGCCAGTGATTGTG | ||
| GIR | 赤霉素调控蛋白 Gibberellin-regulated protein | F:TCCTCTCTTCTCGTTCTT |
| R:GCTCCTCCACAATCAATA | ||
| LRK | 蛋白激酶家族蛋白 Protein kinase family protein | F:CGGAGATGACTGTTGTAA |
| R:TCCAATCAGCAATAATAACG | ||
| CsActin | 内参基因 Actin gene | F:ATTGTTCTCAGTGGTGGTTCTAC |
| R:CCTTTGAGATCCACATCTGCT |
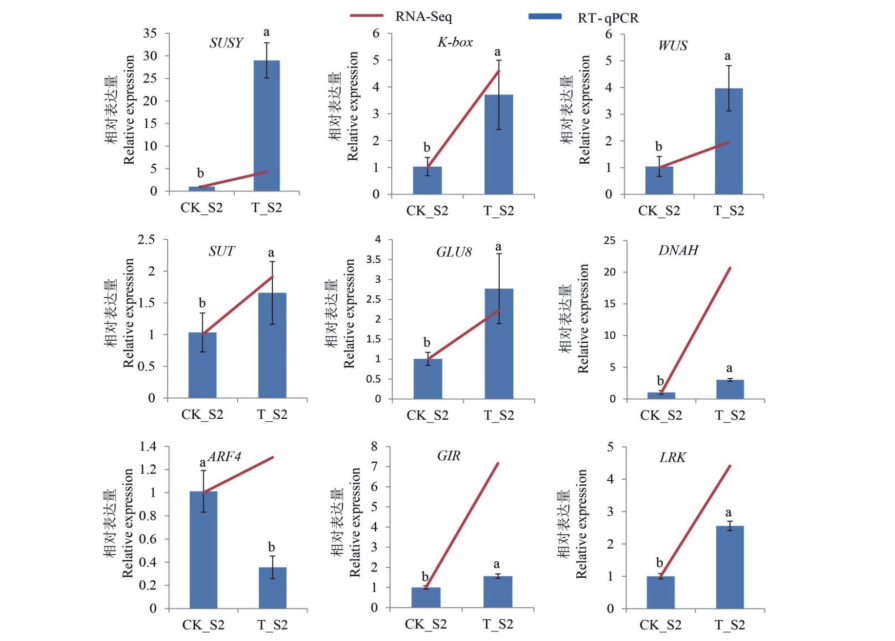
Fig. 6 RT-qPCR analysis of candidate genes Different lowercase letters indicate significant differences in the expression levels of the same gene in diploid and tetraploid cucumber fruits at 0.05 level
| [1] | 潘玉朋, 孟焕文, 陈书霞, 等. 蔬菜作物果形研究进展[J]. 中国蔬菜, 2013(4): 6-13. |
| Pan YP, Meng HW, Chen SX, et al. Research progress on fruit shape of vegetable crops[J]. China Veg, 2013(4): 6-13. | |
| [2] | 黄三文. 黄瓜果实品质性状的基因组学研究[J]. 中国科技成果, 2013(21): 26-28. |
| Hang SW. Genomic studies on fruit quality traits in cucumber[J]. China Sci Technol Achiev, 2013(21): 26-28. | |
| [3] | 王一衡, 黄胜楠, 刘志勇, 等. 大白菜倍性变异引起花发育变化的microRNA调控机制研究[J]. 沈阳农业大学学报, 2018, 49(5): 529-536. |
| Wang YH, Huang SN, Liu ZY, et al. Mechanism of floral development variation resulted from ploidy changes regulated by microRNA in Chinese cabbage[J]. J Shenyang Agric Univ, 2018, 49(5): 529-536. | |
| [4] |
Yin L, Qu JJ, Zhou HW, et al. Comparison of leaf transcriptomes of cassava ‘Xinxuan 048’ diploid and autotetraploid plants[J]. Genes Genomics, 2018, 40(9): 927-935.
doi: 10.1007/s13258-018-0692-2 |
| [5] |
Cheng WW, Tang MJ, Xie Y, et al. Transcriptome-based gene expression profiling of diploid radish(Raphanus sativus L.)and the corresponding autotetraploid[J]. Mol Biol Rep, 2019, 46(1): 933-945.
doi: 10.1007/s11033-018-4549-1 |
| [6] |
Wang HL, Li YQ, Wang SB, et al. Comparative transcriptomic analyses of chlorogenic acid and luteolosides biosynthesis pathways at different flowering stages of diploid and tetraploid Lonicera japonica[J]. PeerJ, 2020, 8: e8690.
doi: 10.7717/peerj.8690 URL |
| [7] | 刘美妍, 周国彦, 李晓丽, 等. 秋水仙素对华北型密刺黄瓜的诱变效应[J]. 中国瓜菜, 2021, 34(11): 10-16. |
| Liu MY, Zhou GY, Li XL, et al. Mutagenesis effect of colchicine on North China cucumber with dense thorny[J]. China Cucurbits Veg, 2021, 34(11): 10-16. | |
| [8] |
Azimian J, Hervan EM, Azadi A, et al. Transcriptome analysis of a Triticum aestivum landrace(Roshan)in response to salt stress conditions[J]. Plant Genet Resour, 2021, 19(3): 261-274.
doi: 10.1017/S1479262121000319 URL |
| [9] |
Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-seq[J]. Nat Methods, 2008, 5(7): 621-628.
doi: 10.1038/nmeth.1226 pmid: 18516045 |
| [10] |
Mereghetti L, Sitkiewicz I, M. Green N, et al. Principal component analysis(PCA)plot showing transcriptome differences between expression microarray data of GBS NEM316 strain incubated in human blood at 37℃[J]. PLoS One, 2009, 4(9): e7145.
doi: 10.1371/journal.pone.0007145 URL |
| [11] | Saetan W, Tian CX, Yu JW, et al. Comparative transcriptome analysis of gill tissue in response to hypoxia in silver Sillago(Sillago sihama)[J]. Animals(Basel), 2020, 10(4): 628. |
| [12] |
Xu YX, Chen JW, Yang ZH, et al. Identification of RNA expression profiles in thyroid cancer to construct a competing endogenous RNA(CeRNA)network of mRNAs, long noncoding RNAs(lncRNAs), and microRNAs(miRNAs)[J]. Med Sci Monit, 2019, 25: 1140-1154.
doi: 10.12659/MSM.912450 URL |
| [13] |
周国彦, 银珊珊, 高佳鑫, 等. 黄瓜AHP基因家族的鉴定及其非生物胁迫表达分析[J]. 生物技术通报, 2022, 38(6): 112-119.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1338 |
| Zhou GY, Yin SS, Gao JX, et al. Identification of AHP gene family in Cucumis sativus and its expression analysis under abiotic stress[J]. Biotechnol Bull, 2022, 38(6): 112-119. | |
| [14] |
Che G, Zhang XL. Molecular basis of cucumber fruit domestication[J]. Curr Opin Plant Biol, 2019, 47: 38-46.
doi: S1369-5266(18)30032-3 pmid: 30253288 |
| [15] |
Weng YQ, Colle M, Wang YH, et al. QTL mapping in multiple populations and development stages reveals dynamic quantitative trait loci for fruit size in cucumbers of different market classes[J]. Theor Appl Genet, 2015, 128(9): 1747-1763.
doi: 10.1007/s00122-015-2544-7 pmid: 26048092 |
| [16] |
Pan YP, Qu SP, Bo KL, et al. QTL mapping of domestication and diversifying selection related traits in round-fruited semi-wild Xishuangbanna cucumber(Cucumis sativus L. var. xishuangbannanesis)[J]. Theor Appl Genet, 2017, 130(7): 1531-1548.
doi: 10.1007/s00122-017-2908-2 URL |
| [17] |
Pan YP, Liang XJ, Gao ML, et al. Round fruit shape in WI7239 cucumber is controlled by two interacting quantitative trait loci with one putatively encoding a tomato SUN homolog[J]. Theor Appl Genet, 2017, 130(3): 573-586.
doi: 10.1007/s00122-016-2836-6 pmid: 27915454 |
| [18] |
Wang LJ, Sheng MY, Wen PC, et al. Morphological, physiological, cytological and phytochemical studies in diploid and colchicine-induced tetraploid plants of Fagopyrum tataricum(L.)Gaertn[J]. Bot Stud, 2017, 58(1): 2.
doi: 10.1186/s40529-016-0157-3 URL |
| [19] | 姜文芝. 不同倍性黄瓜植物学特征及生理生化的研究[D]. 泰安: 山东农业大学, 2017. |
| Jiang WZ. Study on the botanical characteristics and physiological and biochemical of different ploidy cucumber[D]. Tai'an: Shandong Agricultural University, 2017. | |
| [20] | 刘永月, 许建鹏, 李田田, 等. 无毛黄瓜同源四倍体诱导及鉴定[J]. 山东农业科学, 2016, 48(11): 21-25. |
| Liu YY, Xu JP, Li TT, et al. Induction and identification of autotetraploid of glabrous cucumber[J]. Shandong Agric Sci, 2016, 48(11): 21-25. | |
| [21] |
Martin SL, Husband BC. Whole genome duplication affects evolvability of flowering time in an autotetraploid plant[J]. PLoS One, 2012, 7(9): e44784.
doi: 10.1371/journal.pone.0044784 URL |
| [22] | 孙小武, 杨仁喜, 邓大成, 等. 无籽西瓜新品种‘雪峰新二号’的选育[J]. 中国瓜菜, 2018, 31(6): 20-22. |
| Sun XW, Yang RX, Deng DC, et al. Breeding of new seedless watermelon cultivar ‘Xuefeng Xin no. 2’[J]. China Cucurbits Veg, 2018, 31(6): 20-22. | |
| [23] | 高素燕, 焦定量, 商纪鹏, 等. 无籽西瓜新品种‘津蜜55’的选育[J]. 中国瓜菜, 2018, 31(6): 23-25. |
| Gao SY, Jiao DL, Shang JP, et al. Breeding of new seedless watermelon variety ‘Jinmi No.55’[J]. China Cucurbits Veg, 2018, 31(6): 23-25. | |
| [24] |
Adams KL, Cronn R, Percifield R, et al. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing[J]. Proc Natl Acad Sci USA, 2003, 100(8): 4649-4654.
doi: 10.1073/pnas.0630618100 pmid: 12665616 |
| [25] |
Adams KL, Wendel JF. Novel patterns of gene expression in polyploid plants[J]. Trends Genet, 2005, 21(10): 539-543.
pmid: 16098633 |
| [26] |
Alabadí D, Blázquez MA, Carbonell J, et al. Instructive roles for hormones in plant development[J]. Int J Dev Biol, 2009, 53(8/9/10): 1597-1608.
doi: 10.1387/ijdb.072423da URL |
| [27] |
Nemhauser JL, Hong FX, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses[J]. Cell, 2006, 126(3): 467-475.
doi: 10.1016/j.cell.2006.05.050 pmid: 16901781 |
| [28] |
Anfang MR, Shani E. Transport mechanisms of plant hormones[J]. Curr Opin Plant Biol, 2021, 63: 102055.
doi: 10.1016/j.pbi.2021.102055 URL |
| [29] |
Liu JG, Guo SG, He HJ, et al. Dynamic characteristics of sugar accumulation and related enzyme activities in sweet and non-sweet watermelon fruits[J]. Acta Physiol Plant, 2013, 35(11): 3213-3222.
doi: 10.1007/s11738-013-1356-0 URL |
| [30] |
Fan JW, Wang HY, Li X, et al. Down-regulating cucumber sucrose synthase 4(CsSUS4)suppresses the growth and development of flowers and fruits[J]. Plant Cell Physiol, 2019, 60(4): 752-764.
doi: 10.1093/pcp/pcy239 URL |
| [31] |
Baroja-Fernández E, Muñoz FJ, Montero M, et al. Enhancing sucrose synthase activity in transgenic potato(Solanum tuberosum L.)tubers results in increased levels of starch, ADPglucose and UDPglucose and total yield[J]. Plant Cell Physiol, 2009, 50(9): 1651-1662.
doi: 10.1093/pcp/pcp108 pmid: 19608713 |
| [32] |
Xie Y, Ying JL, Xu L, et al. Genome-wide sRNA and mRNA transcriptomic profiling insights into dynamic regulation of taproot thickening in radish(Raphanus sativus L.)[J]. BMC Plant Biol, 2020, 20(1): 373.
doi: 10.1186/s12870-020-02585-z |
| [33] |
D’Aoust MA, Yelle S, Nguyen-Quoc B. Antisense inhibition of tomato fruit sucrose synthase decreases fruit setting and the sucrose unloading capacity of young fruit[J]. Plant Cell, 1999, 11(12): 2407-2418.
doi: 10.1105/tpc.11.12.2407 pmid: 10590167 |
| [34] |
Jiang QY, Hou J, Hao CY, et al. The wheat(T. aestivum)sucrose synthase 2 gene(TaSus2)active in endosperm development is associated with yield traits[J]. Funct Integr Genomics, 2011, 11(1): 49-61.
doi: 10.1007/s10142-010-0188-x URL |
| [1] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [2] | LOU Hui, ZHU Jin-cheng, YANG Yang, ZHANG Wei. Effects of Root Exudates in Resistant and Susceptible Varieties of Cotton on the Growths and Gene Expressions of Fusarium oxysporum [J]. Biotechnology Bulletin, 2023, 39(9): 156-167. |
| [3] | MIAO Yong-mei, MIAO Cui-ping, YU Qing-cai. Properties of Bacillus subtilis Strain BBs-27 Fermentation Broth and the Inhibition of Lipopeptides Against Fusarium culmorum [J]. Biotechnology Bulletin, 2023, 39(9): 255-267. |
| [4] | FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome [J]. Biotechnology Bulletin, 2023, 39(8): 251-261. |
| [5] | CHU Rui, LI Zhao-xuan, ZHANG Xue-qing, YANG Dong-ya, CAO Hang-hang, ZHANG Xue-yan. Screening and Identification of Antagonistic Bacillus spp. Against Cucumber Fusarium wilt and Its Biocontrol Effect [J]. Biotechnology Bulletin, 2023, 39(8): 262-271. |
| [6] | KONG De-zhen, DUAN Zhen-yu, WANG Gang, ZHANG Xin, XI Lin-qiao. Physiological Characteristics and Transcriptome Analysis of Sorghum bicolor × S. Sudanense Seedlings Under Salt-alkali Stress [J]. Biotechnology Bulletin, 2023, 39(6): 199-207. |
| [7] | YANG Yang, ZHU Jin-cheng, LOU Hui, HAN Ze-gang, ZHANG Wei. Transcriptome Analysis of Interaction Between Gossypium barbadense and Fusarium oxysporum f. sp. vasinfectum [J]. Biotechnology Bulletin, 2023, 39(6): 259-273. |
| [8] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [9] | WANG Qi, HU Zhe, FU Wei, LI Guang-zhe, HAO Lin. Regulation of Burkholderia sp. GD17 on the Drought Tolerance of Cucumber Seedlings [J]. Biotechnology Bulletin, 2023, 39(3): 163-175. |
| [10] | HU Li-li, LIN Bo-rong, WANG Hong-hong, CHEN Jian-song, LIAO Jin-ling, ZHUO Kan. Transcriptome and Candidate Effectors Analysis of Pratylenchus brachyurus [J]. Biotechnology Bulletin, 2023, 39(3): 254-266. |
| [11] | YANG Dong-ya, QI Rui-xue LI, Zhao-xuan , LIN Wei, MA Hui, ZHANG Xue-yan. Screening, Identification and Growth-promoting Effect of Antagonistic Bacillus spp. Against Cucumber Fusarium solani [J]. Biotechnology Bulletin, 2023, 39(2): 211-220. |
| [12] | SUN Yan-qiu, XIE Cai-yun, TANG Yue-qin. Construction and Mechanism Analysis of High-temperature Resistant Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2023, 39(11): 226-237. |
| [13] | XU Jun, YE Yu-qing, NIU Ya-jing, HUANG He, ZHANG Meng-meng. Transcriptome Analysis of Rhizome Development in Chrysanthemum× × morifolium [J]. Biotechnology Bulletin, 2023, 39(10): 231-245. |
| [14] | LUO Hao-tian, WANG Long, WANG Yu-qian, WANG Yue, LI Jia-zhen, YANG Meng-ke, ZHANG Jie, DENG Xin, WANG Hong-yan. Genome-wide Identification and Expression Analysis of the RNAi-related Gene Families in Setaria viridis [J]. Biotechnology Bulletin, 2023, 39(1): 175-186. |
| [15] | XIN Jian-pan, LI Yan, ZHAO Chu, TIAN Ru-nan. Transcriptome Sequencing in the Leaves of Pontederia cordata with Cadmium Exposure and Gene Mining in Phenypropanoid Pathways [J]. Biotechnology Bulletin, 2022, 38(6): 198-210. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||