








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (5): 103-111.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1085
Previous Articles Next Articles
LI Xin-yi1,3( ), JIANG Chun-xiu1,3, XUE Li1,3, JIANG Hong-tao1,3, YAO Wei1,3, DENG Zu-hu1,2,3, ZHANG Mu-qing1,3, YU Fan1,3(
), JIANG Chun-xiu1,3, XUE Li1,3, JIANG Hong-tao1,3, YAO Wei1,3, DENG Zu-hu1,2,3, ZHANG Mu-qing1,3, YU Fan1,3( )
)
Received:2022-09-02
Online:2023-05-26
Published:2023-06-08
Contact:
YU Fan
E-mail:lixinyi1776@163.com;yufanky@163.com
LI Xin-yi, JIANG Chun-xiu, XUE Li, JIANG Hong-tao, YAO Wei, DENG Zu-hu, ZHANG Mu-qing, YU Fan. Enhancing Hybridization Signal of Sugarcane Chromosome Oligonucleotide Probe via Multiple Fluorescence Labeled Primers[J]. Biotechnology Bulletin, 2023, 39(5): 103-111.
| 引物名称 Primer name | 引物序列 Primer sequence |
|---|---|
| T7-Cy3 | /iCy3dT/ACGACTCACTATAGGGAGA |
| D9-Cy3 | /iCy3dA/GTGTGTACGGACTAATAAT |
| T7-FAM | /iFAMdT/ACGACTCACTATAGGGAGA |
| D9-FAM | /iFAMdA/GTGTGTACGGACTAATAAT |
| T7-Cy3-3 | /iCy3dT/ACGAC/iCy3dT/CAC/iCy3dT/ATAGGGAGA |
| D9-Cy3-3 | /iCy3dA/G/iCy3dT/G/iCy3dT/GTACGGACTAATAAT |
| T7-FAM-3 | /iFAMdT/ACGAC/iFAMdT/CAC/iFAMdT/ATAGGGAGA |
| D9-FAM-3 | /iFAMdA/G/iFAMdT/G/iFAMdT/GTACGGACTAATAAT |
Table 1 Amplification sequences of oligo probe fluoresc-ence labeled primers
| 引物名称 Primer name | 引物序列 Primer sequence |
|---|---|
| T7-Cy3 | /iCy3dT/ACGACTCACTATAGGGAGA |
| D9-Cy3 | /iCy3dA/GTGTGTACGGACTAATAAT |
| T7-FAM | /iFAMdT/ACGACTCACTATAGGGAGA |
| D9-FAM | /iFAMdA/GTGTGTACGGACTAATAAT |
| T7-Cy3-3 | /iCy3dT/ACGAC/iCy3dT/CAC/iCy3dT/ATAGGGAGA |
| D9-Cy3-3 | /iCy3dA/G/iCy3dT/G/iCy3dT/GTACGGACTAATAAT |
| T7-FAM-3 | /iFAMdT/ACGAC/iFAMdT/CAC/iFAMdT/ATAGGGAGA |
| D9-FAM-3 | /iFAMdA/G/iFAMdT/G/iFAMdT/GTACGGACTAATAAT |
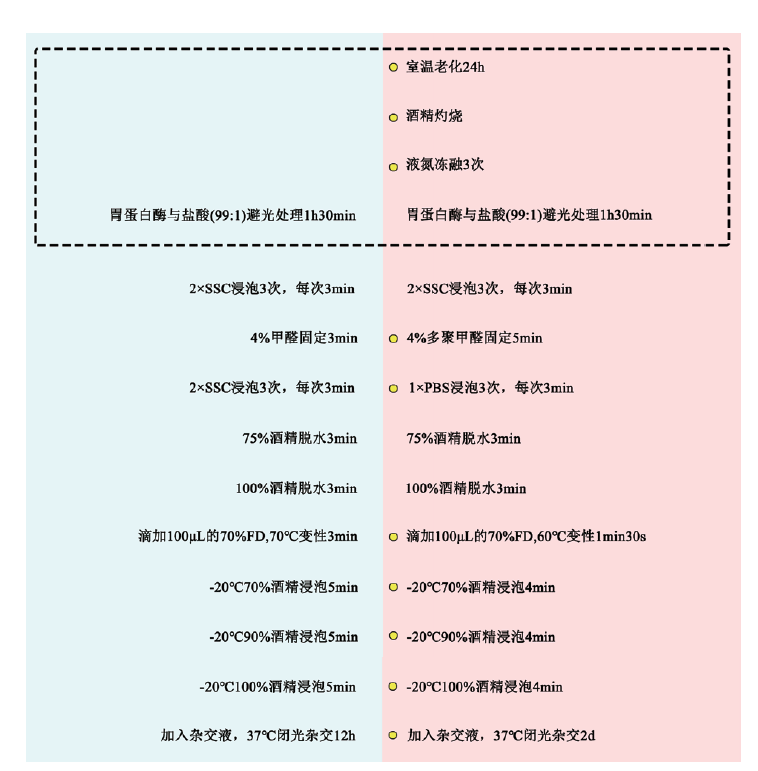
Fig. 2 Comparison of flow before and after optimization of FISH Blue area: Pre-optimization hybridization step; red area: post-optimization hybridization step; yellow marker: improvement step; dashed part: pre-hybridization step area

Fig. 3 Signal capture of Cy3 and FAM with different exposure gradients based on hybridization system optimization a: Imaging of the FAM probe at different exposure duration in the unoptimized hybridization system; b: imaging of the FAM probe at different exposure duration in the optimized hybridization system; c: imaging of the Cy3 probe at different exposure duration in the unoptimized hybridization system; d: imaging of the Cy3 probe at different exposure duration in the optimized hybridization system; e: comparison of fluorescence density before and after optimization system, the red background is marked with Cy3, the green background is marked with FAM.IntDen: Intensity Image. bar=10 μm
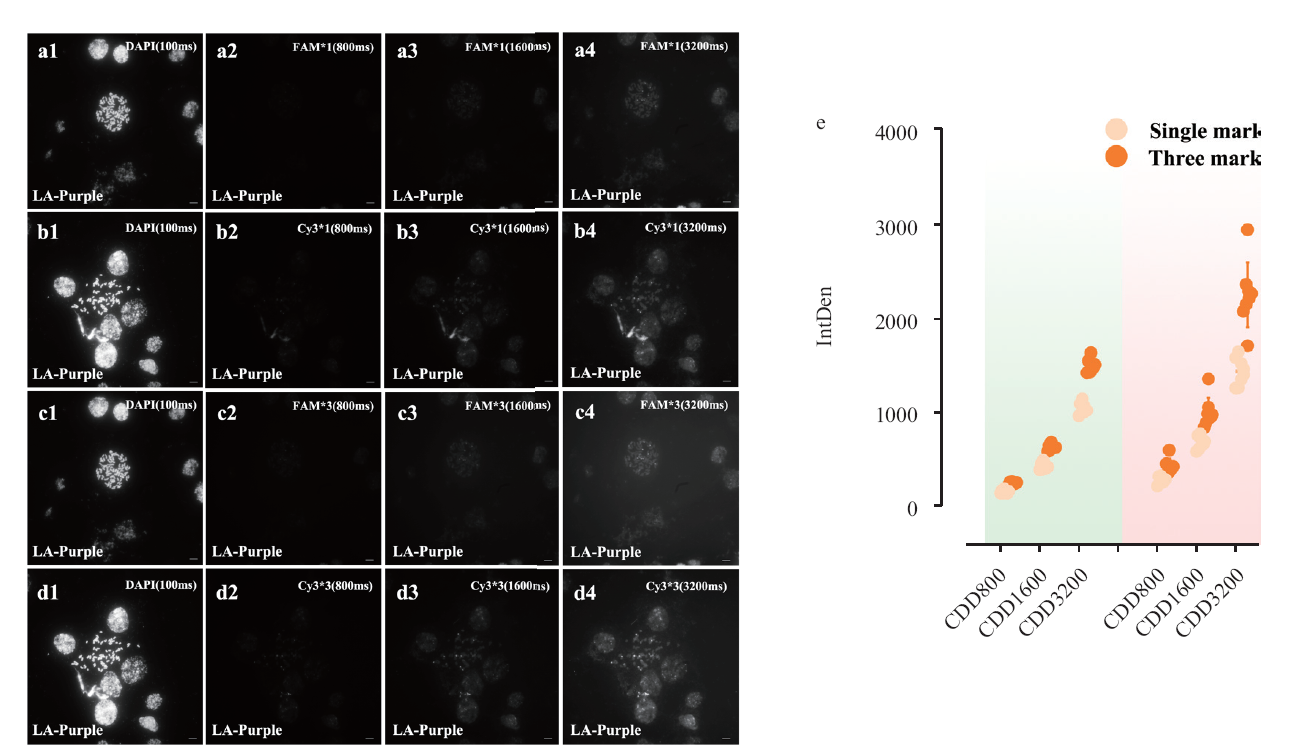
Fig. 4 Signal capture of Cy3 and FAM with different exposure gradients based on different fluorophore group probes a: Imaging of FAM probe at different exposure duration under single fluorescence modification; b: imaging of FAM probe at different exposure duration under three fluorescence modifications; c: imaging of Cy3 probe at different exposure duration under unoptimized hybridization system; d: imaging of Cy3 probe at different exposure duration under three fluorescence modifications; e: comparison graph of fluorescence density under different number of fluorescence modifications, red background for Cy3 marker, green background is FAM marker; IntDen is gray scale value; scale bar is 10 μm
| oligo探针 oligo probes | 高粱染色体 Sorghum chromosome | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| Chr1 | 18 400 | 63 | 120 | 79 | 51 | 84 | 68 | 46 | 55 | 66 | |
| Chr2 | 73 | 12 522 | 55 | 65 | 29 | 55 | 30 | 80 | 34 | 42 | |
| Chr3 | 45 | 46 | 15 771 | 61 | 54 | 47 | 41 | 30 | 125 | 41 | |
| Chr4 | 28 | 47 | 48 | 11 448 | 46 | 42 | 29 | 28 | 18 | 65 | |
| Chr5 | 27 | 34 | 26 | 107 | 3 680 | 29 | 16 | 53 | 25 | 44 | |
| Chr6 | 85 | 20 | 70 | 93 | 48 | 7 959 | 32 | 39 | 41 | 47 | |
| Chr7 | 75 | 81 | 54 | 33 | 40 | 36 | 7 760 | 30 | 39 | 95 | |
| Chr8 | 29 | 34 | 21 | 65 | 143 | 25 | 25 | 5 218 | 24 | 40 | |
| Chr9 | 28 | 39 | 43 | 15 | 25 | 33 | 15 | 26 | 8 592 | 57 | |
| Chr10 | 86 | 112 | 47 | 28 | 46 | 21 | 33 | 60 | 46 | 9 573 | |
Table 2 Number statistics of sorghum genome aligned by S. officinarum oligos probes
| oligo探针 oligo probes | 高粱染色体 Sorghum chromosome | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| Chr1 | 18 400 | 63 | 120 | 79 | 51 | 84 | 68 | 46 | 55 | 66 | |
| Chr2 | 73 | 12 522 | 55 | 65 | 29 | 55 | 30 | 80 | 34 | 42 | |
| Chr3 | 45 | 46 | 15 771 | 61 | 54 | 47 | 41 | 30 | 125 | 41 | |
| Chr4 | 28 | 47 | 48 | 11 448 | 46 | 42 | 29 | 28 | 18 | 65 | |
| Chr5 | 27 | 34 | 26 | 107 | 3 680 | 29 | 16 | 53 | 25 | 44 | |
| Chr6 | 85 | 20 | 70 | 93 | 48 | 7 959 | 32 | 39 | 41 | 47 | |
| Chr7 | 75 | 81 | 54 | 33 | 40 | 36 | 7 760 | 30 | 39 | 95 | |
| Chr8 | 29 | 34 | 21 | 65 | 143 | 25 | 25 | 5 218 | 24 | 40 | |
| Chr9 | 28 | 39 | 43 | 15 | 25 | 33 | 15 | 26 | 8 592 | 57 | |
| Chr10 | 86 | 112 | 47 | 28 | 46 | 21 | 33 | 60 | 46 | 9 573 | |
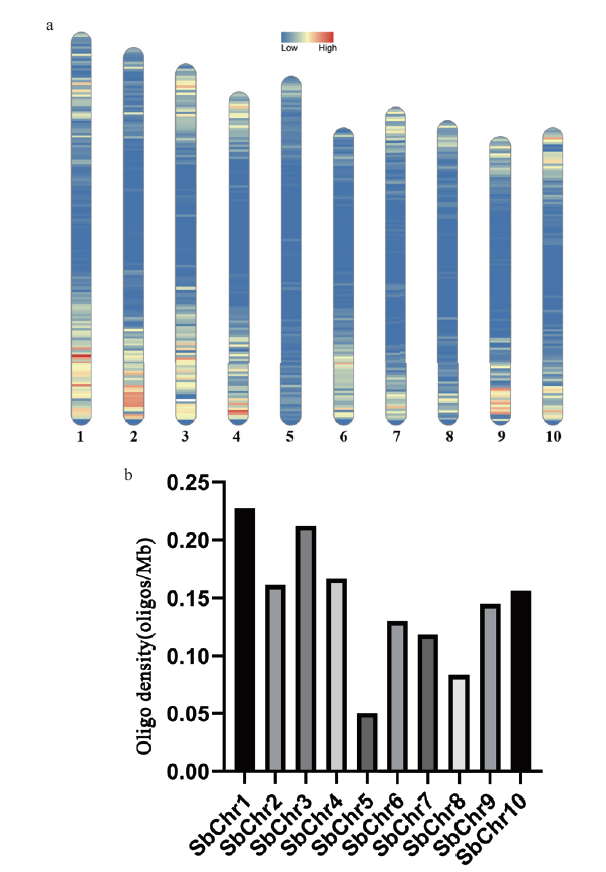
Fig. 5 S. officinarum Oligos sequences distribute on the chromosome of sorghum a: The hot map of Oligo probe distribution. b: The statistics of oligo probe density
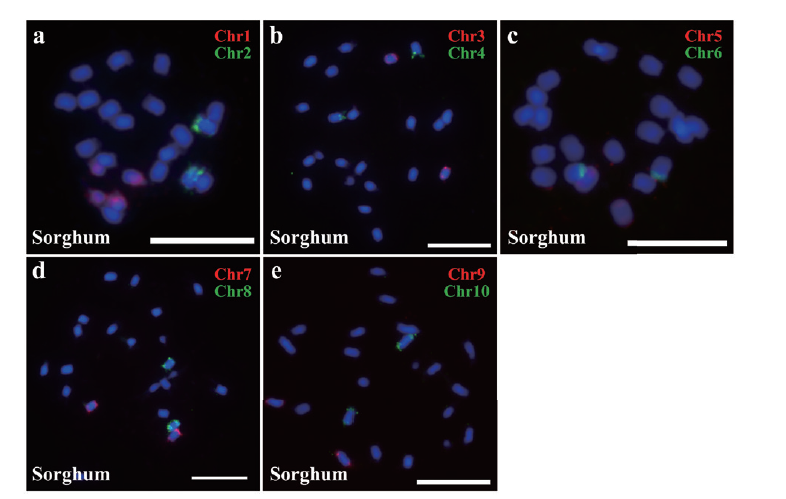
Fig. 6 Oligo probe located on sorghum chromosome a: Probe Chr1 and Chr2 located on sorghum chromosome; b: probe Chr3 and Chr4 located on sorghum chromosome; c: probe Chr5 and Chr6 located on sorghum chromosome; d: probe Chr7 and Chr8 located on sorghum chromosome; e: probe Chr9 and Chr10 located on sorghum chromosome; Cy3 is red, FAM is green; bar=10 μm
| [1] |
Garsmeur O, Droc G, Antonise R, et al. A mosaic monoploid reference sequence for the highly complex genome of sugarcane[J]. Nat Commun, 2018, 9(1): 2638.
doi: 10.1038/s41467-018-05051-5 pmid: 29980662 |
| [2] |
Speicher MR, Gwyn Ballard S, Ward DC. Karyotyping human chromosomes by combinatorial multi-fluor FISH[J]. Nat Genet, 1996, 12(4): 368-375.
pmid: 8630489 |
| [3] |
Xin HY, Zhang T, Wu YF, et al. An extraordinarily stable karyotype of the woody Populus species revealed by chromosome painting[J]. Plant J, 2020, 101(2): 253-264.
doi: 10.1111/tpj.v101.2 URL |
| [4] |
Jiang JM. Fluorescence in situ hybridization in plants: recent developments and future applications[J]. Chromosome Res, 2019, 27(3): 153-165.
doi: 10.1007/s10577-019-09607-z |
| [5] |
Jiang JM, Gill BS. Current status and the future of fluorescence in situ hybridization(FISH)in plant genome research[J]. Genome, 2006, 49(9): 1057-1068.
doi: 10.1139/g06-076 URL |
| [6] |
Liu XY, Sun S, Wu Y, et al. Dual-color oligo-FISH can reveal chromosomal variations and evolution in Oryza species[J]. Plant J, 2020, 101(1): 112-121.
doi: 10.1111/tpj.v101.1 URL |
| [7] | 刘玉玲, 刘震, 李兆国, 等. 棉花oligo-FISH技术建立及其初步应用[J]. 棉花学报, 2017, 29(3): 213-221. |
| Liu YL, Liu Z, Li ZG, et al. Construction and primary application of oligos fluorescence in situ hybridization technology in cotton[J]. Cotton Sci, 2017, 29(3): 213-221. | |
| [8] |
Song XY, Song RR, Zhou JW, et al. Development and application of oligonucleotide-based chromosome painting for chromosome 4D of Triticum aestivum L[J]. Chromosome Res, 2020, 28(2): 171-182.
doi: 10.1007/s10577-020-09627-0 |
| [9] |
Li GR, Zhang T, Yu ZH, et al. An efficient oligo-FISH painting system for revealing chromosome rearrangements and polyploidization in Triticeae[J]. Plant J, 2021, 105(4): 978-993.
doi: 10.1111/tpj.v105.4 URL |
| [10] |
do Vale Martins L, Yu F, Zhao HN, et al. Meiotic crossovers characterized by haplotype-specific chromosome painting in maize[J]. Nat Commun, 2019, 10(1): 4604.
doi: 10.1038/s41467-019-12646-z pmid: 31601818 |
| [11] |
Albert PS, Zhang T, Semrau K, et al. Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships[J]. Proc Natl Acad Sci USA, 2019, 116(5): 1679-1685.
doi: 10.1073/pnas.1813957116 pmid: 30655344 |
| [12] |
Braz GT, Yu F, Zhao HN, et al. Preferential meiotic chromosome pairing among homologous chromosomes with cryptic sequence variation in tetraploid maize[J]. New Phytol, 2021, 229(6): 3294-3302.
doi: 10.1111/nph.17098 pmid: 33222183 |
| [13] |
Yu F, Zhao XW, Chai J, et al. Chromosome-specific painting unveils chromosomal fusions and distinct allopolyploid species in the Saccharum complex[J]. New Phytol, 2022, 233(4): 1953-1965.
doi: 10.1111/nph.v233.4 URL |
| [14] |
Meng Z, Zhang ZL, Yan TY, et al. Comprehensively characterizing the cytological features of Saccharum spontaneum by the development of a complete set of chromosome-specific oligo probes[J]. Front Plant Sci, 2018, 9: 1624.
doi: 10.3389/fpls.2018.01624 URL |
| [15] |
Han YH, Zhang T, Thammapichai P, et al. Chromosome-specific painting in Cucumis species using bulked oligonucleotides[J]. Genetics, 2015, 200(3): 771-779.
doi: 10.1534/genetics.115.177642 URL |
| [16] |
He L, Zhao HN, He J, et al. Extraordinarily conserved chromosomal synteny of Citrus species revealed by chromosome-specific painting[J]. Plant J, 2020, 103(6): 2225-2235.
doi: 10.1111/tpj.v103.6 URL |
| [17] |
Šimoníková D, Němečková A, Karafiátová M, et al. Chromosome painting facilitates anchoring reference genome sequence to chromosomes in situ and integrated karyotyping in banana(Musa spp.)[J]. Front Plant Sci, 2019, 10: 1503.
doi: 10.3389/fpls.2019.01503 pmid: 31824534 |
| [18] |
Yu F, Chai J, Li XT, et al. Chromosomal characterization of Tripidium arundinaceum revealed by oligo-FISH[J]. Int J Mol Sci, 2021, 22(16): 8539.
doi: 10.3390/ijms22168539 URL |
| [19] |
Rauscher JT, Doyle JJ, Brown AHD. Multiple origins and nrDNA internal transcribed spacer homeologue evolution in the Glycine tomentella(Leguminosae)allopolyploid complex[J]. Genetics, 2004, 166(2): 987-998.
pmid: 15020482 |
| [20] | McCarthy EM, Liu JD, Gao LZ, et al. Long terminal repeat retrotransposons of Oryza sativa[J]. Genome Biol, 2002, 3(10): RESEARCH0053. |
| [21] |
Schnable PS, Ware D, Fulton RS, et al. The B73 maize genome: complexity, diversity, and dynamics[J]. Science, 2009, 326(5956): 1112-1115.
doi: 10.1126/science.1178534 pmid: 19965430 |
| [22] |
Zhang JS, Zhang XT, Tang HB, et al. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L[J]. Nat Genet, 2018, 50(11): 1565-1573.
doi: 10.1038/s41588-018-0237-2 |
| [23] | Braz GT, Yu F, do Vale Martins L, et al. Fluorescent in situ hybridization using oligonucleotide-based probes[J]. Methods Mol Biol, 2020, 2148: 71-83. |
| [24] |
Bi YF, Zhao QZ, Yan WK, et al. Flexible chromosome painting based on multiplex PCR of oligonucleotides and its application for comparative chromosome analyses in Cucumis[J]. Plant J, 2020, 102(1): 178-186.
doi: 10.1111/tpj.v102.1 URL |
| [25] |
Zhang T, Liu GQ, Zhao HN, et al. Chorus2: design of genome-scale oligonucleotide-based probes for fluorescence in situ hybridization[J]. Plant Biotechnol J, 2021, 19(10): 1967-1978.
doi: 10.1111/pbi.13610 pmid: 33960617 |
| [26] |
Braz GT, He L, Zhao HN, et al. Comparative oligo-FISH mapping: an efficient and powerful methodology to reveal karyotypic and chromosomal evolution[J]. Genetics, 2018, 208(2): 513-523.
doi: 10.1534/genetics.117.300344 pmid: 29242292 |
| [27] |
Meng Z, Han JL, Lin YJ, et al. Characterization of a Saccharum spontaneum with a basic chromosome number of x=10 provides new insights on genome evolution in genus Saccharum[J]. Theor Appl Genet, 2020, 133(1): 187-199.
doi: 10.1007/s00122-019-03450-w |
| [28] |
Piperidis N, D'Hont A. Sugarcane genome architecture decrypted with chromosome-specific oligo probes[J]. Plant J, 2020, 103(6): 2039-2051.
doi: 10.1111/tpj.v103.6 URL |
| [29] |
Hou LL, Xu M, Zhang T, et al. Chromosome painting and its applications in cultivated and wild rice[J]. BMC Plant Biol, 2018, 18(1): 110.
doi: 10.1186/s12870-018-1325-2 pmid: 29879904 |
| [30] |
Lloyd Evans D, Joshi SV, Wang JP. Whole chloroplast genome and gene locus phylogenies reveal the taxonomic placement and relationship of Tripidium(Panicoideae: Andropogoneae)to sugarcane[J]. BMC Evol Biol, 2019, 19(1): 33.
doi: 10.1186/s12862-019-1356-9 |
| [1] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [2] | CAI Meng-xian, GAO Zuo-min, HU Li-juan, FENG Qun, WANG Hong-cheng, ZHU Bin. Development and Genetic Analysis of Two Nullisomic Lines(NC1 and NC2)in Natural Brassica napus [J]. Biotechnology Bulletin, 2023, 39(3): 81-88. |
| [3] | HUANG Jia-yan, FENG Xiao-yan, SHEN Lin-bo, WANG Wen-zhi, HU Hai-yan, ZHANG Shu-zhen. Cloning of Sugarcane ShPR10 Gene and Study on the Interaction Between ShPR10 Protein and P1 Protein Encoded by Sugarcane Streak Mosaic Virus [J]. Biotechnology Bulletin, 2023, 39(10): 163-174. |
| [4] | GAO Xiao-ning, LIU Rui, WU Zi-lin, WU Jia-yun. Characteristics of Endophytic Fungal and Bacterial Community in the Stalks of Sugarcane Cultivars Resistant to Ratoon Stunting Disease [J]. Biotechnology Bulletin, 2022, 38(6): 166-173. |
| [5] | ZHANG Jing, YOU Chui-huai, CAO Yue, CUI Tian-zhen, YANG Jing-tao, LUO Jun. Sugarcane Rhizosphere Microecology and Its Relationship with Smut Control [J]. Biotechnology Bulletin, 2022, 38(11): 21-31. |
| [6] | FENG Cui-lian, WAN Yue, WANG Jun-gang, FENG Xiao-yan, ZHAO Ting-ting, WANG Wen-zhi, SHEN Lin-bo, ZHANG Shu-zhen. Establishment of a Transformant-specific Detection Method for Cry1Ac-2A-gna Transgenic Sugarcane BCG-17 [J]. Biotechnology Bulletin, 2021, 37(5): 248-258. |
| [7] | FENG Cui-lian, ZHANG Shu-zhen. Breeding of Transgenic Insect-resistant Sugarcane and Strategies for Preventing the Its Resistance to Insects from Loss [J]. Biotechnology Bulletin, 2020, 36(7): 209-219. |
| [8] | GAN Chong-kun, ZHOU Hui-wen, CHEN Rong-fa, FAN Ye-geng, QIU Li-hang, HUANG Xing, LI Yang-rui, LU Xing-gao, WU Jian-ming. Application of Chemical Regulating Technology in Sugarcane Production [J]. Biotechnology Bulletin, 2019, 35(2): 163-170. |
| [9] | ZHANG Bao-qing, SHAO Min, HUANG Yu-xin, HUANG Xing, SONG Xiu-peng, CHEN Hu, WANG Sheng, TAN Qin-liang, YANG Li-tao, LI Yang-rui. Cloning and Expression Analysis of Peroxidase Gene(ScAPX1)from Sugarcane [J]. Biotechnology Bulletin, 2019, 35(12): 31-37. |
| [10] | TANG Shi-yun, YANG Li-tao, LI Yang-rui. Comparative Analysis on Transcriptome Among Different Sugarcane Cultivars Under Low Temperature Stress [J]. Biotechnology Bulletin, 2018, 34(12): 116-124. |
| [11] | ZHAO Ting-ting, WANG Jun-gang, YANG Ben-peng, SHEN Lin-bo, FENG Xiao-yan, WANG Wen-zhi, FENG Cui-lian, XIONG Guo-ru, ZHANG Shu-zhen. The Expression Analysis of Sucrose Transporter Genes in Disease Free Sugarcane [J]. Biotechnology Bulletin, 2018, 34(12): 125-131. |
| [12] | FENG Xiao-yan, WANG Wen-zhi, SHEN Lin-bo ,FENG Cui-lian ,ZHANG Shu-zhen. Research Advances on Sugarcane Streak Mosaic Virus [J]. Biotechnology Bulletin, 2017, 33(7): 22-28. |
| [13] | LI Jia-hui, YUAN Dan, LU Jian-ming, YANG Li-tao, LI Yang-rui ,XING Yong-xiu. Effects of Nitrogen Fertilizer on the Expression of nifH Gene of Endophytic Azotobacter in Sugarcane Leaves [J]. Biotechnology Bulletin, 2017, 33(7): 100-106. |
| [14] | CUI Xue-qiang, ZHANG Shu-zhen, FENG Cui-lian. A Simple Method for Preparing PCR Template of Sugarcane Leaf Tissue [J]. Biotechnology Bulletin, 2016, 32(3): 58-62. |
| [15] | WANG Wen-zhi, YANG Ben-peng, CAI Wen-wei, XIONG Guo-ru, FENG Cui-lian, WANG Jun-gang, WU Yuan-li, SHEN Lin-bo, ZHANG Shu-zhen. On Application of Herbicide Resistant Gene bar and EPSPS in Transgenic Sugarcane [J]. Biotechnology Bulletin, 2016, 32(3): 73-78. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||