








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (5): 112-119.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1247
Previous Articles Next Articles
PAN Guo-qiang1( ), WU Si-yuan1,2, LIU Lu1, GUO Hui-ming1, CHENG Hong-mei1, SU Xiao-feng1(
), WU Si-yuan1,2, LIU Lu1, GUO Hui-ming1, CHENG Hong-mei1, SU Xiao-feng1( )
)
Received:2022-10-10
Online:2023-05-26
Published:2023-06-08
Contact:
SU Xiao-feng
E-mail:15704610584@163.com;suxiaofeng@caas.cn
PAN Guo-qiang, WU Si-yuan, LIU Lu, GUO Hui-ming, CHENG Hong-mei, SU Xiao-feng. Construction and Preliminary Analysis of Verticillim dahliae Mutant Library[J]. Biotechnology Bulletin, 2023, 39(5): 112-119.
| 引物名称Primer name | 序列 Sequence(5'-3') |
|---|---|
| VdrDNA-qPCR-F | CCGCCGGTCCATCAGTCTCTCTGTTTATAC |
| VdrDNA-qPCR-R | CGCCTGCGGGACTCCGATGCGAGCTGTAAC |
| NbActin-qPCR-F | GGCTTCCTCAAGGTCGGCTATG |
| NbActin-qPCR-R | GCTGCATGTCATCCCACTTCTTC |
Table 1 Primer sequences for detection of biomass of V. dahliae in Nicotiana benthamiana root by qPCR
| 引物名称Primer name | 序列 Sequence(5'-3') |
|---|---|
| VdrDNA-qPCR-F | CCGCCGGTCCATCAGTCTCTCTGTTTATAC |
| VdrDNA-qPCR-R | CGCCTGCGGGACTCCGATGCGAGCTGTAAC |
| NbActin-qPCR-F | GGCTTCCTCAAGGTCGGCTATG |
| NbActin-qPCR-R | GCTGCATGTCATCCCACTTCTTC |
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| AP1 | Provided by Takara Genome Walking Kit, unknown sequence |
| SP1 | ATCGTTGGTGTCGATGTCAGCTCC |
| SP2 | GCGTTTCGGGTTTACCTCTTCCAG |
| SP3 | CGAGATCAAGCAGATCAACGGTCG |
Table 2 Primer sequences for the amplification of flanking sequences at insertion site
| 引物名称 Primer name | 序列 Sequence(5'-3') |
|---|---|
| AP1 | Provided by Takara Genome Walking Kit, unknown sequence |
| SP1 | ATCGTTGGTGTCGATGTCAGCTCC |
| SP2 | GCGTTTCGGGTTTACCTCTTCCAG |
| SP3 | CGAGATCAAGCAGATCAACGGTCG |
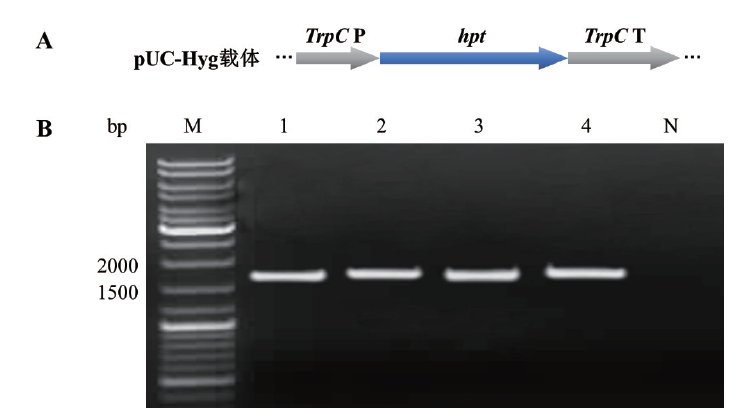
Fig. 1 Schematic diagram and PCR amplification of hygromycin resistance gene expression cassette in pUC-Hyg A: Schematic diagram of hygromycin resistance gene expression cassette in pUC-Hyg. B: PCR amplification of hygromycin resistance gene expression cassette fragment. M: DNA marker. 1-4: Hygromycin resistance gene expression cassette fragment. N: Negative control(ddH2O)

Fig. 2 PCR identification of randomly inserted mutants of V. dahlia M: DNA marker. 1-5: PCR identification products of transformant 1-5 genomic DNA. WT: PCR identification products of V991 genomic DNA. N: Negative control(ddH2O)

Fig. 3 Colony growth phenotype of randomly inserted mutants on PDA and different carbon source medium A: Colony morphology of V. dahliae V991 and 5 randomly inserted mutants were cultured on PDA and different carbon source medium for 12 d. B: Colony diameters of V. dahliae V991 and 5 randomly inserted mutants were cultured on PDA and different carbon source medium for 12 d. C: Spore production of V. dahliae V991 and 5 randomly inserted mutants cultured on PDA and different carbon source medium for 12 d. Error bars indicate the standard deviations and different letters indicate significant differences at P<0.05
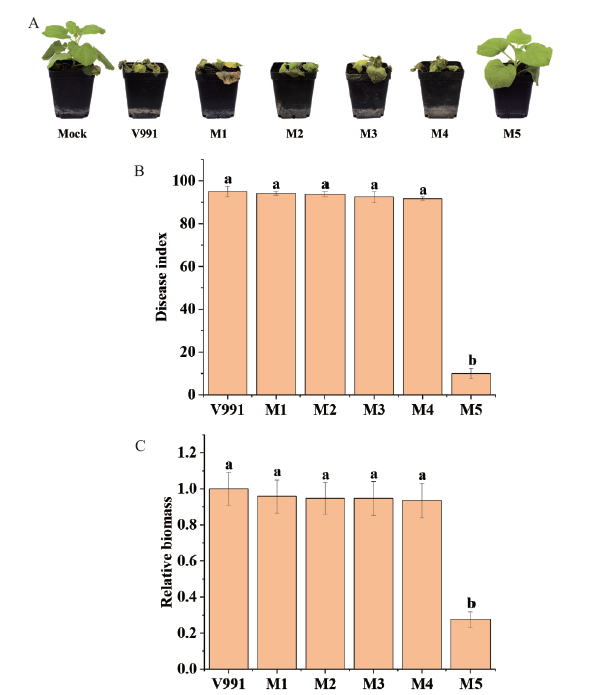
Fig. 4 Pathogenicity of randomly inserted mutants of V. dahliae to N. benthamiana A: Disease incidence of N. benthamiana plants after inoculation with V. dahliae V991 and 5 randomly inserted mutants for 12 d. Mock: N. benthamiana plants that were not inoculated with V. dahliae. B: Disease index of N. benthamiana plants after inoculation with V. dahliae V991 and 5 randomly inserted mutants for 12 d. C: Fungal biomass in the root tissues of N. benthamiana after inoculation with V. dahliae V991 and 5 randomly inserted mutants for 12 d. Error bars indicate the standard deviations and different letters indicate significant differences at P<0.05
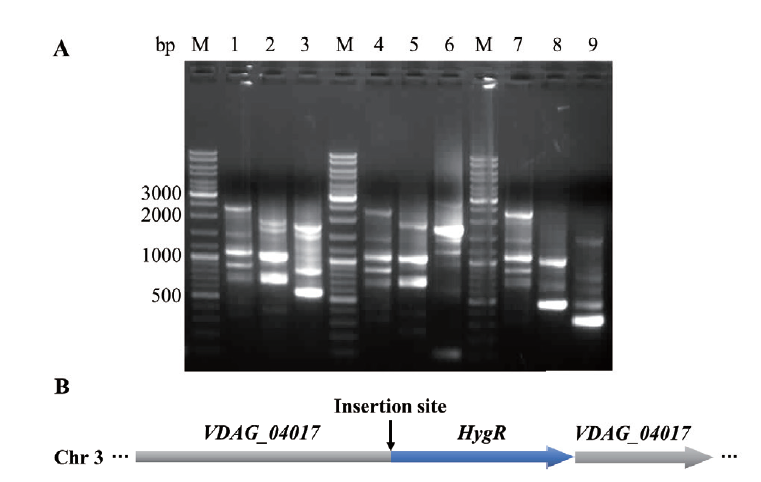
Fig. 5 Flanking sequence analysis of insertion site for randomly inserted mutant M5 A: Flanking sequence detection of the insertion site of hygromycin resistance gene expression cassette fragment(M: DNA marker. 1-9: three rounds of PCR reaction amplification products). B: Schematic diagram of insertion site of hygromycin resistance gene expression cassette fragment
| [1] |
Ali F, Qanmber G, Li YH, et al. Genome-wide identification of Gossypium INDETERMINATE DOMAIN genes and their expression profiles in ovule development and abiotic stress responses[J]. J Cotton Res, 2019, 2: 3-18.
doi: 10.1186/s42397-019-0021-6 |
| [2] |
Wendel JF. New world tetraploid cottons contain old world cytoplasm[J]. Proc Natl Acad Sci USA, 1989, 86(11): 4132-4136.
pmid: 16594050 |
| [3] |
Shaban M, Miao YH, Ullah A, et al. physiological and molecular mechanism of defense in cotton against Verticillium dahliae[J]. Plant Physiol Biochem, 2018, 125: 193-204.
doi: 10.1016/j.plaphy.2018.02.011 URL |
| [4] |
Daayf F. Verticillium wilts in crop plants: Pathogen invasion and host defence responses[J]. Can J Plant Pathol, 2015, 37(1): 8-20.
doi: 10.1080/07060661.2014.989908 URL |
| [5] |
Gui YJ, Chen JY, Zhang DD, et al. Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate-binding module 1[J]. Environ Microbiol, 2017, 19(5): 1914-1932.
doi: 10.1111/1462-2920.13695 URL |
| [6] |
Zhang Y, Gao YH, Wang HL, et al. Verticillium dahliae secretory effector PevD1 induces leaf senescence by promoting ORE1-mediated ethylene biosynthesis[J]. Mol Plant, 2021, 14(11): 1901-1917.
doi: 10.1016/j.molp.2021.07.014 pmid: 34303024 |
| [7] |
Su XF, Lu GQ, Li XK, et al. Host-induced gene silencing of an adenylate kinase gene involved in fungal energy metabolism improves plant resistance to Verticillium dahliae[J]. Biomolecules, 2020, 10(1): 127-142.
doi: 10.3390/biom10010127 URL |
| [8] |
Zhang T, Jin Y, Zhao JH, et al. Host-induced gene silencing of the target gene in fungal cells confers effective resistance to the cotton wilt disease pathogen Verticillium dahliae[J]. Mol Plant, 2016, 9(6): 939-942.
doi: 10.1016/j.molp.2016.02.008 pmid: 26925819 |
| [9] |
Klimes A, Dobinson KF. A hydrophobin gene, VDH1, is involved in microsclerotial development and spore viability in the plant pathogen Verticillium dahliae[J]. Fungal Genet Biol, 2006, 43(4): 283-294.
doi: 10.1016/j.fgb.2005.12.006 pmid: 16488633 |
| [10] |
Chi MH, Park SY, Kim S, et al. A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host[J]. PLoS Pathog, 2009, 5(4): e1000401-e1000416.
doi: 10.1371/journal.ppat.1000401 URL |
| [11] |
Chou TH, Tzean SS. Protoplasting, regeneration and transformation of medicinal mushroom Ganoderma multipileum using succinate dehydrogenase mutation gene as a selection marker[J]. Ann Microbiol, 2016, 66(1): 111-120.
doi: 10.1007/s13213-015-1087-0 URL |
| [12] |
Rehman L, Su XF, Guo HM, et al. Protoplast transformation as a potential platform for exploring gene function in Verticillium dahliae[J]. BMC Biotechnol, 2016, 16(1): 57-65.
doi: 10.1186/s12896-016-0287-4 URL |
| [13] |
Su XF, Rehman L, Guo HM, et al. The oligosaccharyl transferase subunit STT3 mediates fungal development and is required for virulence in Verticillium dahliae[J]. Curr Genet, 2018, 64(1): 235-246.
doi: 10.1007/s00294-017-0729-0 URL |
| [14] |
Wei F. Effects of individual and combined use of bio-fumigation-derived products on the viability of Verticillium dahliae microsclerotia in soil[J]. Crop Prot, 2016, 79: 170-176.
doi: 10.1016/j.cropro.2015.09.008 URL |
| [15] |
Faino L, de Jonge R, Thomma BPHJ. The transcriptome of Verticillium dahliae-infected Nicotiana benthamiana determined by deep RNA sequencing[J]. Plant Signal Behav, 2012, 7(9): 1065-1069.
doi: 10.4161/psb.21014 URL |
| [16] |
Klosterman SJ, Subbarao KV, Kang S, et al. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens[J]. PLoS Pathog, 2011, 7(7): e1002137-e1002155.
doi: 10.1371/journal.ppat.1002137 URL |
| [17] |
Dobinson KF, Grant SJ, Kang S. Cloning and targeted disruption, via Agrobacterium tumefaciens-mediated transformation, of a trypsin protease gene from the vascular wilt fungus Verticillium dahliae[J]. Curr Genet, 2004, 45(2): 104-110.
doi: 10.1007/s00294-003-0464-6 URL |
| [18] |
Mullins ED, Chen X, Romaine P, et al. Agrobacterium-mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer[J]. Phytopathology, 2001, 91(2): 173-180.
doi: 10.1094/PHYTO.2001.91.2.173 pmid: 18944391 |
| [19] |
Amey RC. PEG-mediated and Agrobacterium-mediated transformation in the mycopathogen Verticillium fungicola[J]. Mycol Res, 2002, 106(1): 4-11.
doi: 10.1017/S0953756201005251 URL |
| [20] |
Dobrowolska A, Staczek P. Development of transformation system for Trichophyton rubrum by electroporation of germinated conidia[J]. Curr Genet, 2009, 55(5): 537-542.
doi: 10.1007/s00294-009-0264-8 pmid: 19629488 |
| [21] |
Wang D, Chen JY, Song J, et al. Cytotoxic function of xylanase VdXyn4 in the plant vascular wilt pathogen Verticillium dahliae[J]. Plant Physiol, 2021, 187(1): 409-429.
doi: 10.1093/plphys/kiab274 pmid: 34618145 |
| [22] |
Zhang J, Zhang YY, Yang JF, et al. The α-1, 6-mannosyltransferase VdOCH1 plays a major role in microsclerotium formation and virulence in the soil-borne pathogen Verticillium dahliae[J]. Fungal Biol, 2019, 123(7): 539-546.
doi: 10.1016/j.funbio.2019.05.007 URL |
| [23] |
Gao F, Zhou BJ, Li GY, et al. A glutamic acid-rich protein identified in Verticillium dahliae from an insertional mutagenesis affects microsclerotial formation and pathogenicity[J]. PLoS One, 2010, 5(12): e15319-e15328.
doi: 10.1371/journal.pone.0015319 URL |
| [24] |
Zhang DD, Wang XY, Chen JY, et al. Identification and characterization of a pathogenicity-related gene VdCYP1 from Verticillium dahliae[J]. Sci Rep, 2016, 6: 27979-27990.
doi: 10.1038/srep27979 |
| [25] |
Maruthachalam K, Klosterman SJ, Kang S, et al. Identification of pathogenicity-related genes in the vascular wilt fungus Verticillium dahliae by Agrobacterium tumefaciens-mediated T-DNA insertional mutagenesis[J]. Mol Biotechnol, 2011, 49(3): 209-221.
doi: 10.1007/s12033-011-9392-8 pmid: 21424547 |
| [26] |
Yang C, Liu R, Pang JH, et al. Poaceae-specific cell wall-derived oligosaccharides activate plant immunity via OsCERK1 during Magnaporthe oryzae infection in rice[J]. Nat Commun, 2021, 12(1): 2178-2190.
doi: 10.1038/s41467-021-22456-x |
| [1] | LIU Li-hui, CHU Jin-hua, SUI Yu-xin, CHEN Yang, CHENG Gu-yue. Research Progress of Main Virulence Factors in Salmonella [J]. Biotechnology Bulletin, 2022, 38(9): 72-83. |
| [2] | ZHAO Jing-ya, PENG Meng-ya, ZHANG Shi-yu, SHAN Yi-xuan, XING Xiao-ping, SHI Yan, LI Hai-yang, YANG Xue, LI Hong-lian, CHEN Lin-lin. Role of C2H2 Zinc Finger Transcription Factor FpCzf7 in the Growth and Pathogenicity of Fusarium pseudograminearum [J]. Biotechnology Bulletin, 2022, 38(8): 216-224. |
| [3] | CHEN Fu-nuan, HUANG Yu, CAI Jia, WANG Zhong-liang, JIAN Ji-chang, WANG Bei. Structure of ABC Transporter and Research Progress of It in Bacterial Pathogenicity [J]. Biotechnology Bulletin, 2022, 38(6): 43-52. |
| [4] | TIAN Li, LI Jun-jiao, DAI Xiao-feng, ZHANG Dan-dan, CHEN Jie-yin. From Functional Genes to Biological Characteristics:The Molecular Basis of Pathogenicity in Verticillium dahliae [J]. Biotechnology Bulletin, 2022, 38(1): 51-69. |
| [5] | LIU Sha-yu, CAO Jian, LI Meng, LIU Zhi-qiang, LI Xiao-yu. Biological Function of a Zn2Cys6 Transcription Factor CgAswA in Colletotrichum gloeosporioides [J]. Biotechnology Bulletin, 2021, 37(9): 161-170. |
| [6] | LUO Li-li, ZHANG Hao, YANG Mei-xin, WANG Yun-fei, XU Jing-sheng, XU Jin, YAO Qiang, FENG Jie. Temperature-related Pathogenicity Differentiation of Wheat Head Blight in Huang and Huai River Valleys and Northeast China Wheat Regions [J]. Biotechnology Bulletin, 2021, 37(4): 47-55. |
| [7] | ZHANG Wen-ze, ZHANG Yan-Li, MEN Yan-ming, ZHANG Yu-jiao, SUN Zhi-xin, LI Wen-hui, LU Guo-dong, QI Yao-yao. Functional Analysis of Arginine N-methyltransferase Gene MoHMT1 in Magnaporthe oryzae [J]. Biotechnology Bulletin, 2019, 35(12): 38-44. |
| [8] | ZHANG Meng-tian, PEI Juan, LI Guo ,ZHAO Hui ,CHEN Jian-quan, ZHU Jian-bo ,WANG Ai-ying. Identification and Pathogenicity Analysis of Cotton Verticillium Wilt from Shihezi Region of Xinjiang [J]. Biotechnology Bulletin, 2018, 34(6): 73-78. |
| [9] | GUO Yun-feng, AN Bang. Functional Analysis of NADPH Oxidases in Colletotrichum gloeosporioides from Hevea brasiliensis [J]. Biotechnology Bulletin, 2018, 34(10): 165-171. |
| [10] | LI Yang, LI He, ZHOU Guo-ying, LIU Jun-ang. Identification of a New Anthracnose Pathogen Colletotrichum camelliae and Its Pathogenicity Test on Camellia oleifera [J]. Biotechnology Bulletin, 2016, 32(6): 96-102. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||