








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (8): 272-282.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0038
Previous Articles Next Articles
FANG Lan1( ), LI Yan-yan2, JIANG Jian-wei1, CHENG Sheng1, SUN Zheng-xiang1(
), LI Yan-yan2, JIANG Jian-wei1, CHENG Sheng1, SUN Zheng-xiang1( ), ZHOU Yi1(
), ZHOU Yi1( )
)
Received:2023-01-17
Online:2023-08-26
Published:2023-09-05
Contact:
SUN Zheng-xiang, ZHOU Yi
E-mail:1605867934@qq.com;sunzhengxiang9904@126.com;zhouyi@yangtzeu.edu.cn
FANG Lan, LI Yan-yan, JIANG Jian-wei, CHENG Sheng, SUN Zheng-xiang, ZHOU Yi. Isolation, Identification and Growth-promoting Characteristics of Endohyphal Bacterium 7-2H from Endophytic Fungi of Spiranthes sinensis[J]. Biotechnology Bulletin, 2023, 39(8): 272-282.
| 检测基因 Detected gene | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 预变性 Initial denaturation | 变性Denaturation | 退火Annealing | 延伸Extension | 最终延伸 Final extension |
|---|---|---|---|---|---|---|---|
| 30 cycles | |||||||
| 16S rDNA | 27F | AGAGTTTGATCCTGGCTCAG | 94℃, 2 min | 94℃, 30 s | 55℃, 30 s | 72℃, 90 s | 72℃, 5 min |
| 1492R | TACGGCTACCTTGTTACGACTT | ||||||
| ChvE | ChvE-F101 | GCTGGTCTTCCTTGTAGTAA | 56℃, 30 s | 72℃, 60 s | |||
| ChvE-R653 | AACGCCTTCTTCTTCTATGA | ||||||
Table 1 PCR amplification procedure
| 检测基因 Detected gene | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 预变性 Initial denaturation | 变性Denaturation | 退火Annealing | 延伸Extension | 最终延伸 Final extension |
|---|---|---|---|---|---|---|---|
| 30 cycles | |||||||
| 16S rDNA | 27F | AGAGTTTGATCCTGGCTCAG | 94℃, 2 min | 94℃, 30 s | 55℃, 30 s | 72℃, 90 s | 72℃, 5 min |
| 1492R | TACGGCTACCTTGTTACGACTT | ||||||
| ChvE | ChvE-F101 | GCTGGTCTTCCTTGTAGTAA | 56℃, 30 s | 72℃, 60 s | |||
| ChvE-R653 | AACGCCTTCTTCTTCTATGA | ||||||
| 试验名称 Test name | 结果 Result | 试验名称 Test name | 结果 Result | |
|---|---|---|---|---|
| 淀粉水解Starch hydrolysis | - | 葡萄糖 Glucose | + | |
| 纤维素降解 Cellulose degradation | - | 乳糖 Lactose | + | |
| 蛋白降解 Protein degradation | - | 甘露醇 Mannitol | + | |
| 几丁质降解 Chitin degradation | - | 接触酶 Catalase | + | |
| 硝酸盐还原 Nitrate reduction | + | 甲基红 Methyl red | - |
Table 2 Physiological and biochemical determination of 7-2H
| 试验名称 Test name | 结果 Result | 试验名称 Test name | 结果 Result | |
|---|---|---|---|---|
| 淀粉水解Starch hydrolysis | - | 葡萄糖 Glucose | + | |
| 纤维素降解 Cellulose degradation | - | 乳糖 Lactose | + | |
| 蛋白降解 Protein degradation | - | 甘露醇 Mannitol | + | |
| 几丁质降解 Chitin degradation | - | 接触酶 Catalase | + | |
| 硝酸盐还原 Nitrate reduction | + | 甲基红 Methyl red | - |
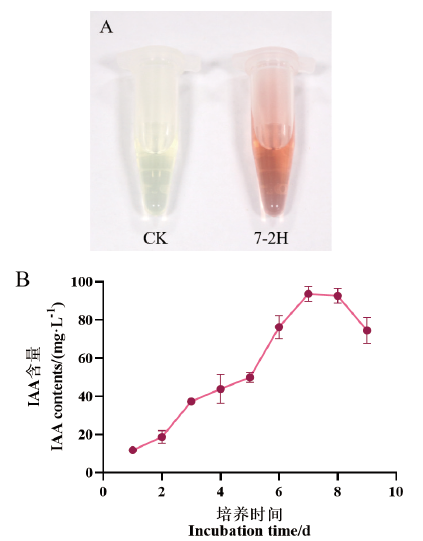
Fig. 5 Determination of IAA-producing ability of strain 7-2H A: Qualitative detection of IAA-producing ability of strain 7-2H. B: IAA contents by strain 7-2H at different culture time
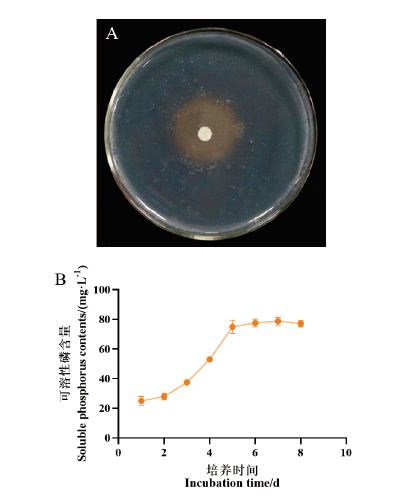
Fig. 6 Determination of siderophore-producing ability of strain 7-2H A: Effect of strain 7-2H growing on CAS detection plate for 5 d. B: Siderophores relative contents by strain 7-2H at different culture time
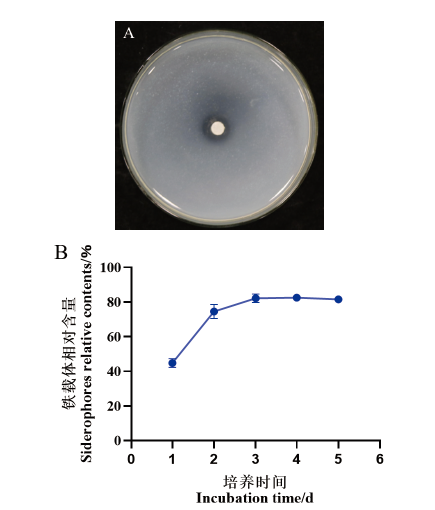
Fig. 7 Determination of phosphate-solubilizing ability of strain 7-2H A: Effect of strain 7-2H on NBRIP inorganic phosphorus plate for 3 d. B: Soluble phosphorus contents by strain 7-2H at different culture time

Fig. 8 Effects of strain 7-2H on the growth indexes of rice seed seedlings A: Shoot length. B: Root length. C: Fresh weight. D: Dry weight. E: Growth situation. ****: Extremely significant difference(P<0.01); *: significant difference(P<0.05)
| 数据库 Database | 基因数量 Gene number | 基因注释比例 Gene annotation ratio/% |
|---|---|---|
| Uniprot | 3 081 | 58.12 |
| Pfam | 4 501 | 84.91 |
| Refseq | 5 087 | 95.96 |
| Nr | 3 014 | 56.86 |
| Tigrfam | 5 127 | 96.72 |
| GO | 2 961 | 55.86 |
| KEGG | 2 663 | 50.24 |
| COG | 2 107 | 39.75 |
Table 3 Database distribution of gene functional annotation from the strain 7-2H
| 数据库 Database | 基因数量 Gene number | 基因注释比例 Gene annotation ratio/% |
|---|---|---|
| Uniprot | 3 081 | 58.12 |
| Pfam | 4 501 | 84.91 |
| Refseq | 5 087 | 95.96 |
| Nr | 3 014 | 56.86 |
| Tigrfam | 5 127 | 96.72 |
| GO | 2 961 | 55.86 |
| KEGG | 2 663 | 50.24 |
| COG | 2 107 | 39.75 |
| [1] | 国家环境保护总局. 中国珍稀濒危保护植物名录[M]. 北京: 科学出版社, 1987: 23-28. |
| State Environmental Protection Administration. List of rare and endangered plants in China[M]. Beijing: Science Press, 1987: 23-28. | |
| [2] | 高越, 郭顺星, 邢晓科. 兰科植物种子共生萌发真菌多样性及共生萌发机制研究进展[J]. 菌物学报, 2019, 38(11): 1808-1825. |
| Gao Y, Guo SX, Xing XK. Fungal diversity and mechanisms of symbiotic germination of orchid seeds: a review[J]. Mycosystema, 2019, 38(11): 1808-1825. | |
| [3] | 裴东方, 吴秋秋, 莫俊鑫, 等. 盘龙参内生真菌多样性及其拮抗作用研究[J]. 江西农业大学学报, 2019, 41(2): 365-371. |
| Pei DF, Wu QQ, Mo JX, et al. Diversity of endophytic fungi from Spiranthes sinensis(pers.) Ames and their antifungal activity[J]. Acta Agric Univ Jiangxiensis, 2019, 41(2): 365-371. | |
| [4] |
Braga RM, Padilla G, Araújo WL. The biotechnological potential of Epicoccum spp.: diversity of secondary metabolites[J]. Crit Rev Microbiol, 2018, 44(6): 759-778.
doi: 10.1080/1040841X.2018.1514364 URL |
| [5] |
Koné Y, Alves E, Silveira PR, et al. Control of blast disease caused by Pyricularia oryzae with Epicoccum nigrum and microscopic studies of their interaction with rice plants under greenhouse conditions[J]. Biol Control, 2022, 167: 104840.
doi: 10.1016/j.biocontrol.2022.104840 URL |
| [6] |
Jin ZX, Li DD, Liu TY, et al. Cultural endophytic fungi associated with Dendrobium officinale: identification, diversity estimation and their antimicrobial potential[J]. Curr Sci, 2017, 112(8): 1690.
doi: 10.18520/cs/v112/i08/1690-1697 URL |
| [7] |
Ali Aslani M, Harighi B, Abdollahzadeh J. Screening of endofungal bacteria isolated from wild growing mushrooms as potential biological control agents against brown blotch and internal stipe necrosis diseases of Agaricus bisporus[J]. Biol Control, 2018, 119: 20-26.
doi: 10.1016/j.biocontrol.2018.01.006 URL |
| [8] |
Desirò A, Faccio A, Kaech A, et al. Endogone, one of the oldest plant-associated fungi, host unique Mollicutes-related endobacteria[J]. New Phytol, 2015, 205(4): 1464-1472.
doi: 10.1111/nph.13136 pmid: 25345989 |
| [9] |
Naumann M, Schüssler A, Bonfante P. The obligate endobacteria of arbuscular mycorrhizal fungi are ancient heritable components related to the Mollicutes[J]. ISME J, 2010, 4(7): 862-871.
doi: 10.1038/ismej.2010.21 pmid: 20237515 |
| [10] | Pakvaz S, Soltani J. Endohyphal bacteria from fungal endophytes of the Mediterranean cypress(Cupressus sempervirens)exhibit in vitro bioactivity[J]. For Pathol, 2016, 46(6): 569-581. |
| [11] |
Shaffer JP, Zalamea PC, Sarmiento C, et al. Context-dependent and variable effects of endohyphal bacteria on interactions between fungi and seeds[J]. Fungal Ecol, 2018, 36: 117-127.
doi: 10.1016/j.funeco.2018.08.008 URL |
| [12] | 刘泽, 孙翔, 刘晓玲, 等. 真菌内共生细菌研究进展[J]. 菌物学报, 2019, 38(10): 1581-1599. |
| Liu Z, Sun X, Liu XL, et al. Research progress on endofungal bacteria[J]. Mycosystema, 2019, 38(10): 1581-1599. | |
| [13] |
Ruiz-Herrera J, León-Ramírez C, Vera-Nuñez A, et al. A novel intracellular nitrogen-fixing symbiosis made by Ustilago maydis and Bacillus spp[J]. New Phytol, 2015, 207(3): 769-777.
doi: 10.1111/nph.13359 pmid: 25754368 |
| [14] |
Almendras K, García J, Carú M, et al. Nitrogen-fixing bacteria associated with Peltigera cyanolichens and Cladonia chloroli-chens[J]. Molecules, 2018, 23(12): 3077.
doi: 10.3390/molecules23123077 URL |
| [15] |
Shaffer JP, U'Ren JM, Gallery RE, et al. An endohyphal bacterium(Chitinophaga, bacteroidetes)alters carbon source use by Fusarium Keratoplasticum(F. Solani species complex, Nectriaceae)[J]. Front Microbiol, 2017, 8: 350.
doi: 10.3389/fmicb.2017.00350 pmid: 28382021 |
| [16] |
Lackner G, Hertweck C. Impact of endofungal bacteria on infection biology, food safety, and drug development[J]. PLoS Pathog, 2011, 7(6): e1002096.
doi: 10.1371/journal.ppat.1002096 URL |
| [17] |
Lumini E, Bianciotto V, Jargeat P, et al. Presymbiotic growth and sporal morphology are affected in the arbuscular mycorrhizal fungus Gigaspora margarita cured of its endobacteria[J]. Cell Microbiol, 2007, 9(7): 1716-1729.
pmid: 17331157 |
| [18] |
Hoffman MT, Gunatilaka MK, Wijeratne K, et al. Endohyphal bacterium enhances production of indole-3-acetic acid by a foliar fungal endophyte[J]. PLoS One, 2013, 8(9): e73132.
doi: 10.1371/journal.pone.0073132 URL |
| [19] |
Cheng S, Jiang JW, Tan LT, et al. Plant growth-promoting ability of mycorrhizal Fusarium strain KB-3 enhanced by its IAA producing endohyphal bacterium, Klebsiella aerogenes[J]. Front Microbiol, 2022, 13: 855399.
doi: 10.3389/fmicb.2022.855399 URL |
| [20] |
Partida-Martinez LP, Hertweck C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production[J]. Nature, 2005, 437(7060): 884-888.
doi: 10.1038/nature03997 |
| [21] |
Jiang JW, Zhang K, Cheng S, et al. Fusarium oxysporum KB-3 from Bletilla striata: an orchid mycorrhizal fungus[J]. Mycorrhiza, 2019, 29(5): 531-540.
doi: 10.1007/s00572-019-00904-3 |
| [22] | 裴东方. 盘龙参内生菌多样性及一株内生细菌的抑菌活性研究[D]. 荆州: 长江大学, 2020. |
| Pei DF. Diversity of endophytes from Spiranthes sinensis and the antifungal activity of one endophytic bacterium[D]. Jingzhou: Yangtze University, 2020. | |
| [23] | 王亚军, 冯炬威, 李雅倩, 等. 高产铁载体菌Burkholderia vietnamiensis YQ9促生特性研究及其对重金属胁迫条件下种子萌发的影响[J]. 环境科学学报, 2022, 42(2): 430-437. |
| Wang YJ, Feng JW, Li YQ, et al. Studies on growth-promoting properties of an efficient siderophore producing bacterium, Burkholderia vietnamiensis YQ9, and its effects on seed germination under heavy metal stress[J]. Acta Sci Circumstantiae, 2022, 42(2): 430-437. | |
| [24] |
Hoffman MT, Arnold AE. Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes[J]. Appl Environ Microbiol, 2010, 76(12): 4063-4075.
doi: 10.1128/AEM.02928-09 URL |
| [25] | 谭利涛, 成胜, 苏杭, 等. 真菌内生蜡样芽孢杆菌7-1Y的分离鉴定及其功能探索[J]. 江西农业大学学报, 2022, 44(4): 900-909. |
| Tan LT, Cheng S, Su H, et al. Isolation, identification and functional exploration of endohyphal Bacillus cereus 7-1Y[J]. Acta Agric Univ Jiangxiensis, 2022, 44(4): 900-909. | |
| [26] | 谢家仪, 董光军, 刘振英. 扫描电镜的微生物样品制备方法[J]. 电子显微学报, 2005, 24(4): 440. |
| Xie JY, Dong GJ, Liu ZY. Preparation method of microbial samples by scanning electron microscope[J]. J Chin Electron Microsc Soc, 2005, 24(4): 440. | |
| [27] | 东秀珠, 蔡妙英. 常见细菌系统鉴定手册[M]. 北京: 科学出版社, 2001: 62-65. |
| Dong XZ, Cai MY. Handbook of identification of common bacterial systems[M]. Beijing: Science Press, 2001: 62-65. | |
| [28] |
He FL, Nair GR, Soto CS, et al. Molecular basis of ChvE function in sugar binding, sugar utilization, and virulence in Agrobacterium tumefaciens[J]. J Bacteriol, 2009, 191(18): 5802-5813.
doi: 10.1128/JB.00451-09 URL |
| [29] | 罗兴, 邹兰, 吴清山, 等. 乌头产吲哚乙酸内生细菌遗传多样性、抗逆性及其对水稻幼苗生长的影响[J]. 微生物学报, 2022, 62(4): 1485-1500. |
| Luo X, Zou L, Wu QS, et al. Genetic diversity, stress resistance, and effect on rice seedling growth of indoleacetic acid-producing endophytic bacteria isolated from Aconitum carmichaelii Debeaux[J]. Acta Microbiol Sin, 2022, 62(4): 1485-1500. | |
| [30] | 杨常娥, 鲁艳莉, 倪捍成, 等. 创伤弧菌产铁载体菌株的筛选及其诱导条件的响应面优化[J]. 食品工业科技, 2017, 38(3): 159-165. |
| Yang CE, Lu YL, Ni HC, et al. Screening of siderophore-producing Vibrio vulnificus and optimization of induction conditions using response surface methodology[J]. Sci Technol Food Ind, 2017, 38(3): 159-165. | |
| [31] |
钱婷, 叶建仁. 巨大芽孢杆菌ZS-3溶无机磷机制及其对樟树的促生作用[J]. 生物技术通报, 2020, 36(8): 45-52.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0114 |
| Qian T, Ye JR. The mechanism of dissolving inorganic phosphorus by Bacillus megaterium ZS-3 and its growth promotion of Cinnamomum camphora[J]. Biotechnol Bull, 2020, 36(8): 45-52. | |
| [32] | Gholamalizadeh R, Khodakaramian G, Ebadi AA. Assessment of rice associated bacterial ability to enhance rice seed germination and rice growth promotion[J]. Braz Arch Biol Technol, 2017, 60: e17160410. |
| [33] |
姚延轩, 接伟光, 杜燕, 等. 根瘤菌的分类、鉴定及应用技术研究现状[J]. 中国农学通报, 2020, 36(15): 100-105.
doi: 10.11924/j.issn.1000-6850.casb19010151 |
| Yao YX, Jie WG, Du Y, et al. Taxonomy, identification and application of Rhizobium[J]. Chin Agric Sci Bull, 2020, 36(15): 100-105. | |
| [34] | Saghafi D, Ghorbanpour M, Lajayer BA. Efficiency of Rhizobium strains as plant growth promoting rhizobacteria on morpho-physiological properties of Brassica napus L. under salinity stress[J]. J Soil Sci Plant Nutr, 2018(ahead). |
| [35] |
Zhao JJ, Zhao X, Wang JR, et al. Isolation, identification and characterization of endophytic bacterium Rhizobium oryzihabitans sp. nov., from rice root with biotechnological potential in agriculture[J]. Microorganisms, 2020, 8(4): 608.
doi: 10.3390/microorganisms8040608 URL |
| [36] |
Agnolucci M, Battini F, Cristani C, et al. Diverse bacterial communities are recruited on spores of different arbuscular mycorrhizal fungal isolates[J]. Biol Fertil Soils, 2015, 51(3): 379-389.
doi: 10.1007/s00374-014-0989-5 URL |
| [37] | Shaffer JP, Sarmiento C, Zalamea PC, et al. Diversity, specificity, and phylogenetic relationships of endohyphal bacteria in fungi that inhabit tropical seeds and leaves[J]. Front Ecol Evol, 2016, 4: 116. |
| [38] |
Sharma M, Schmid M, Rothballer M, et al. Detection and identification of bacteria intimately associated with fungi of the order Sebacinales[J]. Cell Microbiol, 2008, 10(11): 2235-2246.
doi: 10.1111/j.1462-5822.2008.01202.x pmid: 18637023 |
| [39] | Guo HJ, Glaeser SP, Alabid I, et al. The abundance of endofungal bacterium Rhizobium radiobacter(syn. Agrobacterium tumefaciens)increases in its fungal host piriformospora indica during the tripartite sebacinalean symbiosis with higher plants[J]. Front Microbiol, 2017, 8: 629. |
| [40] | Swarnalakshmi K, Yadav V, Tyagi D, et al. Significance of plant growth promoting rhizobacteria in grain legumes: growth promotion and crop production[J]. Plants: Basel, 2020, 9(11): E1596. |
| [41] |
Saghafi D, Ghorbanpour M, Ajirloo HS, et al. Enhancement of growth and salt tolerance in Brassica napus L. seedlings by halotolerant Rhizobium strains containing ACC-deaminase activity[J]. Plant Physiol Rep, 2019, 24(2): 225-235.
doi: 10.1007/s40502-019-00444-0 |
| [42] |
Chaudhary T, Gera R, Shukla P. Deciphering the potential of Rhizobium pusense MB-17a, a plant growth-promoting root endophyte, and functional annotation of the genes involved in the metabolic pathway[J]. Front Bioeng Biotechnol, 2021, 8: 617034.
doi: 10.3389/fbioe.2020.617034 URL |
| [43] |
Ghignone S, Salvioli A, Anca I, et al. The genome of the obligate endobacterium of an AM fungus reveals an interphylum network of nutritional interactions[J]. ISME J, 2012, 6(1): 136-145.
doi: 10.1038/ismej.2011.110 pmid: 21866182 |
| [44] |
Li Q, Li XL, Chen C, et al. Analysis of bacterial diversity and communities associated with Tricholoma matsutake fruiting bodies by barcoded pyrosequencing in Sichuan Province, southwest China[J]. J Microbiol Biotechnol, 2016, 26(1): 89-98.
doi: 10.4014/jmb.1505.05008 URL |
| [1] | WANG Teng-hui, GE Wen-dong, LUO Ya-fang, FAN Zhen-yu, WANG Yu-shu. Gene Mapping of Kale White Leaves Based on Whole Genome Re-sequencing of Extreme Mixed Pool(BSA) [J]. Biotechnology Bulletin, 2023, 39(9): 176-182. |
| [2] | GUO Shao-hua, MAO Hui-li, LIU Zheng-quan, FU Mei-yuan, ZHAO Ping-yuan, MA Wen-bo, LI Xu-dong, GUAN Jian-yi. Whole Genome Sequencing and Comparative Genome Analysis of a Fish-derived Pathogenic Aeromonas Hydrophila Strain XDMG [J]. Biotechnology Bulletin, 2023, 39(8): 291-306. |
| [3] | ZHANG Zhi-xia, LI Tian-pei, ZENG Hong, ZHU Xi-xian, YANG Tian-xiong, MA Si-nan, HUANG Lei. Genome Sequencing and Bioinformatics Analysis of Gelidibacter sp. PG-2 [J]. Biotechnology Bulletin, 2023, 39(3): 290-300. |
| [4] | HE Meng-ying, LIU Wen-bin, LIN Zhen-ming, LI Er-tong, WANG Jie, JIN Xiao-bao. Whole Genome Sequencing and Analysis of an Anti Gram-positive Bacterium Gordonia WA4-43 [J]. Biotechnology Bulletin, 2023, 39(2): 232-242. |
| [5] | ZHANG Ao-jie, LI Qing-yun, SONG Wen-hong, YAN Shao-hui, TANG Ai-xing, LIU You-yan. Whole Genome Sequencing Analysis of a Phenol-degrading Strain Alcaligenes faecalis JF101 [J]. Biotechnology Bulletin, 2023, 39(10): 292-303. |
| [6] | WANG Shuai, LV Hong-rui, ZHANG Hao, WU Zhan-wen, XIAO Cui-hong, SUN Dong-mei. Whole-Genome Sequencing Identification of Phosphate-solubilizing Bacteria PSB-R and Analysis of Its Phosphate-solubilizing Properties [J]. Biotechnology Bulletin, 2023, 39(1): 274-283. |
| [7] | WEN Chang, LIU Chen, LU Shi-yun, XU Zhong-bing, AI Chao-fan, LIAO Han-peng, ZHOU Shun-gui. Biological Characteristics and Genome Analysis of a Novel Multidrug-resistant Shigella flexneri Phage [J]. Biotechnology Bulletin, 2022, 38(9): 127-135. |
| [8] | LI Ji-hong, JING Yu-ling, MA Gui-zhen, GUO Rong-jun, LI Shi-dong. Genome Construction of Achromobacter 77 and Its Characteristics on Chemotaxis and Antibiotic Resistance [J]. Biotechnology Bulletin, 2022, 38(9): 136-146. |
| [9] | ZHANG Ze-ying, FAN Qing-feng, DENG Yun-feng, WEI Ting-zhou, ZHOU Zheng-fu, ZHOU Jian, WANG Jin, JIANG Shi-jie. Whole Genome Sequencing and Comparative Genomic Analysis of a High-yield Lipase-producing Strain WCO-9 [J]. Biotechnology Bulletin, 2022, 38(10): 216-225. |
| [10] | CHEN Ti-qiang, XU Xiao-lan, SHI Lin-chun, ZHONG Li-Yi. Sequencing and Analysis of the Whole Genome of Zizhi Cultivar ‘Wuzhi No.2’(Ganoderma sp. strain Zizhi S2) [J]. Biotechnology Bulletin, 2021, 37(11): 42-56. |
| [11] | GUO He-bao, WANG Xing, HE Shan-wen, ZHANG Xiao-xia. Phenotypic Characteristics Combined with Genomic Analysis to Identify Different Colony Morphology Bacillus velezensis ACCC 19742 [J]. Biotechnology Bulletin, 2020, 36(2): 142-148. |
| [12] | LIANG Han-qiao, Dilidaer Kudereti, BAI Fei-rong, MIAO Lei, LI Nan-nan, LIU Su-hui, CHEN Jian-guo, LIU Yang, SUN Yu-ping. Isolation and Identification of Endophytic Fungi from the Rhizomes of Xinjiang-specific Plant Brassica rapa [J]. Biotechnology Bulletin, 2017, 33(2): 131-136. |
| [13] | ZHOU Zhen-yu, HU Jin-li, SU Xin, LIU Dong-xue, BU Ning, MA Lian-ju. Identification and Resistance of an Endophytic Fungus YD02 Strain in Wild Glycine soja [J]. Biotechnology Bulletin, 2017, 33(11): 106-111. |
| [14] | Wang Xingwen,Wang Jiaqi,Zhao Shengguo,Li Fadi,Bu Dengpan. The Application of Uncultured Methods in the Study of Ruminal Methanogen Population [J]. Biotechnology Bulletin, 2014, 0(6): 67-74. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||