








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (8): 283-290.doi: 10.13560/j.cnki.biotech.bull.1985
Previous Articles Next Articles
RAO Zi-huan( ), XIE Zhi-xiong(
), XIE Zhi-xiong( )
)
Received:2023-01-09
Online:2023-08-26
Published:2023-09-05
Contact:
XIE Zhi-xiong
E-mail:RZHmio@whu.edu.cn;zxxie@whu.edu.cn
RAO Zi-huan, XIE Zhi-xiong. Isolation and Identification of a Cellulose-degrading Strain of Olivibacter jilunii and Analysis of Its Degradability[J]. Biotechnology Bulletin, 2023, 39(8): 283-290.
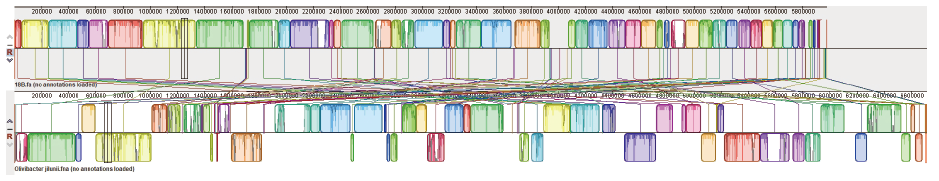
Fig. 4 Genome-wide covariance alignment of strain 18B and O. jilunii 14-2AT It in the upper part of the figure it is strain 18B and in the lower part is O. jilunii 14-2AT

Fig. 5 Genome alignment within the genus Olivibacter Complete genome sequence information of related strains in Olivibacter genus was downloaded from NCBI and compared with strain 18B. In the figure, sitiens indicates the strain O. sitiensis AW-6T, domesticus indicates O. domesticus DC186T, and LS_1 indicates O. sp.LS-1. jilunii refers to strain O. jilunii 14-2AT
| 项目 Item | 18B | O. jilunii 14-2AT |
|---|---|---|
| 生长温度/℃ | 12-48 | 4-42 |
| 生长pH | 6.0-8.0 | 6.0-9.0 |
| NaCl耐受/% | 0-6 | 0-5 |
| 氧化酶 | + | - |
| 硝酸盐还原 | + | + |
| 糖酵解 | - | + |
| VP | + | + |
| β-半乳糖苷酶 | + | + |
| 革兰氏染色 | - | - |
| MacConkey agar | +(Red) | +(Red) |
Table 1 Comparison of common physiological and bioche-mical characteristics between 18B and O. jilunii 14-2AT
| 项目 Item | 18B | O. jilunii 14-2AT |
|---|---|---|
| 生长温度/℃ | 12-48 | 4-42 |
| 生长pH | 6.0-8.0 | 6.0-9.0 |
| NaCl耐受/% | 0-6 | 0-5 |
| 氧化酶 | + | - |
| 硝酸盐还原 | + | + |
| 糖酵解 | - | + |
| VP | + | + |
| β-半乳糖苷酶 | + | + |
| 革兰氏染色 | - | - |
| MacConkey agar | +(Red) | +(Red) |
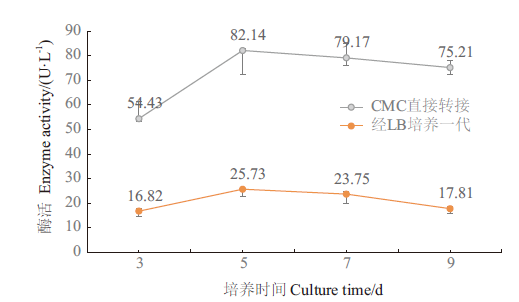
Fig. 6 Variation of 18B cellulase activity with incubation time The gray line is the cellulase activity of the strain after transfer from CMC-Na culture environment, and the red line is the cellulase activity of the strain after transfer from CMC-Na culture environment to LB culture for one generation. The enzyme activity of O. jilunii 14-2AT was not detected
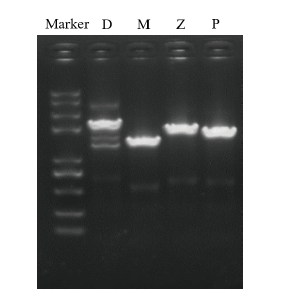
Fig. 7 Gene amplification after induction of expression Marker is Trans2k plusII. D is the precursor gene of endoglucanase D, the length is 1 812 bp. M is cellulase M gene with length of 1 119 bp. Z is the precursor gene of endoglucanase Z, the length of which is 1 542 bp. P is a pure endoglucanase gene with a length of 1 395 bp
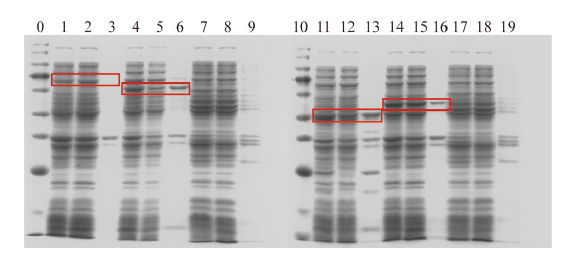
Fig. 8 Protein assay of cellulase gene induced expression 0: Maker. 1: The whole solution broken after D protein expression; 2: the supernatant broken after D protein expression; 3: the precipitation broken after D protein expression; 4: the whole fluid broken after Z protein expression; 5: the supernatant broken after Z protein expression; 6: the precipitation broken after Z protein expression; 7: the whole solution of cell breaking after no-load induction expression; 8: the supernatant of cell breaking after no-load induction expression; 9: the precipitation of cell breaking after no-load induction expression; 10: marker; 11: the whole solution broken after M protein expression; 12: the supernatant broken after M protein expression; 13: the precipitation broken after M protein expression; 14: the whole fluid broken after P protein expression; 15: the supernatant broken after P protein expression; 16: the precipitation broken after P protein expression; 17: the whole solution of cell breaking after no-load induction expression; 18: the supernatant of cell breaking after no-load induction expression; 19: the precipitation of cell breaking after no-load induction expression. The red box marked the target protein region
| [1] |
Arevalo-Gallegos A, Ahmad Z, Asgher M, et al. Lignocellulose: a sustainable material to produce value-added products with a zero waste approach-a review[J]. Int J Biol Macromol, 2017, 99: 308-318.
doi: S0141-8130(16)32991-9 pmid: 28254573 |
| [2] |
Sharma A, Tewari R, Rana SS, et al. Cellulases: classification, methods of determination and industrial applications[J]. Appl Biochem Biotechnol, 2016, 179(8): 1346-1380.
doi: 10.1007/s12010-016-2070-3 pmid: 27068832 |
| [3] |
Djuric Ilic D, Dotzauer E, Trygg L, et al. Introduction of large-scale biofuel production in a district heating system-an opportunity for reduction of global greenhouse gas emissions[J]. J Clean Prod, 2014, 64: 552-561.
doi: 10.1016/j.jclepro.2013.08.029 URL |
| [4] |
Sadhu S. Cellulase production by bacteria: a review[J]. Br Microbiol Res J, 2013, 3(3): 235-258.
doi: 10.9734/BMRJ URL |
| [5] | Juturu V, Wu JC. Microbial cellulases: engineering, production and applications[J]. Renew Sustain Energy Rev, 2014, 33: 188-203. |
| [6] |
Ntougias S, Fasseas C, Zervakis GI. Olivibacter sitiensis Gen. nov., sp. nov., isolated from alkaline olive-oil mill wastes in the region of Sitia, Crete[J]. Int J Syst Evol Microbiol, 2007, 57(Pt 2): 398-404.
doi: 10.1099/ijs.0.64561-0 URL |
| [7] | Lane DJ. 16S/23S rRNA sequencing[M]//Stackebrandt E, Goodfellow M, eds. Nucleic Acid Techniques in Bacterial Systematics. Chichester, UK: J. Wiley & Sons, 1991: 115-175. |
| [8] |
Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Mol Biol Evol, 2018, 35(6): 1547-1549.
doi: 10.1093/molbev/msy096 pmid: 29722887 |
| [9] |
Sokal RR. Numerical taxonomy[J]. Sci Am, 1966, 215(6): 106-116.
doi: 10.1038/scientificamerican1266-106 URL |
| [10] | Swofford DL. PAUP* 4.0b10: phylogenetic analysis using parsimony(and other methods)[Software]. 2003. |
| [11] |
Darling ACE, Mau B, Blattner FR, et al. Mauve: multiple alignment of conserved genomic sequence with rearrangements[J]. Genome Res, 2004, 14(7): 1394-1403.
doi: 10.1101/gr.2289704 pmid: 15231754 |
| [12] |
Alikhan NF, Petty NK, Ben Zakour NL, et al. BLAST Ring Image Generator(BRIG): simple prokaryote genome comparisons[J]. BMC Genomics, 2011, 12: 402.
doi: 10.1186/1471-2164-12-402 |
| [13] |
Chen K, Tang SK, Wang GL, et al. Olivibacter jilunii sp. nov., isolated from DDT-contaminated soil[J]. Int J Syst Evol Microbiol, 2013, 63(Pt 3): 1083-1088.
doi: 10.1099/ijs.0.042416-0 URL |
| [14] |
Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar[J]. Anal Chem, 1959, 31(3): 426-428.
doi: 10.1021/ac60147a030 URL |
| [15] |
Wang WK, Liang CM. Enhancing the compost maturation of swine manure and rice straw by applying bioaugmentation[J]. Sci Rep, 2021, 11(1): 6103.
doi: 10.1038/s41598-021-85615-6 |
| [16] |
Konovalova A, Kahne DE, Silhavy TJ. Outer membrane biogenesis[J]. Annu Rev Microbiol, 2017, 71: 539-556.
doi: 10.1146/annurev-micro-090816-093754 pmid: 28886680 |
| [17] |
Ang SK, Shaza EM, Adibah Y, et al. Production of cellulases and xylanase by Aspergillus fumigatus SK1 using untreated oil palm trunk through solid state fermentation[J]. Process Biochem, 2013, 48(9): 1293-1302.
doi: 10.1016/j.procbio.2013.06.019 URL |
| [18] |
Gilkes NR, Kilburn DG, Miller RC Jr, et al. Bacterial cellulases[J]. Bioresour Technol, 1991, 36(1): 21-35.
doi: 10.1016/0960-8524(91)90097-4 URL |
| [19] | 章沙沙, 徐健峰, 柳增善. 纤维素降解菌的研究与应用进展[J]. 工业微生物, 2021, 51(2): 46-52. |
| Zhang SS, Xu JF, Liu ZS. Progress in research and application of cellulose-degrading microorganisms[J]. Ind Microbiol, 2021, 51(2): 46-52. | |
| [20] |
Niyonzima FN. Detergent-compatible bacterial cellulases[J]. J Basic Microbiol, 2019, 59(2): 134-147.
doi: 10.1002/jobm.v59.2 URL |
| [1] | YOU Zi-juan, CHEN Han-lin, DENG Fu-cai. Research Progress in the Extraction and Functional Activities of Bioactive Peptides from Fish Skin [J]. Biotechnology Bulletin, 2023, 39(7): 91-104. |
| [2] | CHE Yong-mei, GUO Yan-ping, LIU Guang-chao, YE Qing, LI Ya-hua, ZHAO Fang-gui, LIU Xin. Isolation and Identification of Bacterial Strain C8 and B4 and Their Halotolerant Growth-promoting Effects and Mechanisms [J]. Biotechnology Bulletin, 2023, 39(5): 276-285. |
| [3] | WANG Feng-ting, WANG Yan, SUN Ying, CUI Wen-jing, QIAO Kai-bin, PAN Hong-yu, LIU Jin-liang. Isolation and Identification of Saline-alkali Tolerant Aspergillus terreus SYAT-1 and Its Activities Against Plant Pathogenic Fungi [J]. Biotechnology Bulletin, 2023, 39(2): 203-210. |
| [4] | ZU Xue, ZHOU Hu, ZHU Hua-jun, REN Zuo-hua, LIU Er-ming. Isolation and Identification of Bacillus subtilis K-268 and Its Biological Control Effect on Rice Blast [J]. Biotechnology Bulletin, 2022, 38(6): 136-146. |
| [5] | WANG Chun-yan, LA Gui-xiao, SU Xiu-hong, LI Meng, DONG Cheng-ming. Screening of Endophytic Bacteria from Rehmannia glutinosa at Different Stages and Analysis of Their Growth-promoting Characteristics [J]. Biotechnology Bulletin, 2022, 38(4): 242-252. |
| [6] | ZHANG Gong-you, WANG Yi-han, GUO Min, ZHANG Ting-ting, WANG Bing, LIU Hong-mei. Isolation and Identification of a Cellulase-producing Endophytic Fungus in Paris polyphylla var. yunnanensis [J]. Biotechnology Bulletin, 2022, 38(2): 95-104. |
| [7] | NIU Hong-yu, SHU Qian, YANG Hai-jun, YAN Zhi-yong, TAN Ju. Isolation, Identification, Degradation Characteristics and Metabolic Pathway of an Efficient Sodium Dodecyl Sulfate-degrading Bacterium [J]. Biotechnology Bulletin, 2022, 38(12): 287-299. |
| [8] | CUI Xin-yu, LI Rong-rong, CAI Rui, WANG Yan, ZHENG Meng-hu, XU Chun-cheng. Isolation,Identification of Lactic Acid Degrading Bacteria in Alfalfa Silage and Their Degradation Characterization [J]. Biotechnology Bulletin, 2021, 37(9): 58-67. |
| [9] | WEI Xiao-bo, HOU Ying, CHENG Hao-jie, QIN Cui-li, NIU Ming-fu, XU Jian-qiang. Isolation,Identification of Phenol-degrading Pseudoxanthomonas sp. BF-6 and Its Degradation Characteristics and Pathway [J]. Biotechnology Bulletin, 2021, 37(10): 72-80. |
| [10] | LIN Jia-ming, GE Hui, LIN Ke-bing, YANG Zhang-wu, ZHOU Chen, WU Jian-shao, WANG Guo-dong, ZHANG Zhe, YANG Qiu-hua, WANG Yi-lei. Isolation,Identification and Antibiotic Sensitivity Analysis of Bacterial Pathogen from Litopenaeus vannamei with Black Gill Disease [J]. Biotechnology Bulletin, 2020, 36(8): 120-128. |
| [11] | LI Lin-chao, ZHANG Chao, DONG Qing, GUO Cheng, ZHOU Bo, GAO Zheng. Isolation and Identification of Cellulose Degrading Microorganisms in Composting Process [J]. Biotechnology Bulletin, 2019, 35(9): 165-171. |
| [12] | WANG Xue-han, MA Qiang, TIAN Yuan, HU Jing, LIU Hui-rong. Cultivable Myxobacteria and Their Antibiotic Activities in the Hulun Buir Area of Inner Mongolia [J]. Biotechnology Bulletin, 2019, 35(9): 224-233. |
| [13] | JIN Hong-jie, CAO Hong, LIU Hong, ZHENG Shuang, JIANG Chao. Isolation of Endophytic Fungi from Cinnamomum camphora Leaves,Screening and Identification of Biologically Active Strains [J]. Biotechnology Bulletin, 2019, 35(3): 53-58. |
| [14] | LIU Wan, ZHAO Shan-shan ,JIAO Zhen ,YANG Feng-yuan ,WANG Yan-ping. Screening Cellulase-producing Strains and Ion Beam Mutagenesis on Them [J]. Biotechnology Bulletin, 2018, 34(6): 149-154. |
| [15] | LI Jing, XIE Jin, LI Xiang-nan, ZHOU Zhi-fan, LIU Feng-lu, CHEN Yu-hui. Isolation,Identification and Antimicrobial Activity of Mycoparasites(Pestalotiopsis)from Aecidium pourthiaea [J]. Biotechnology Bulletin, 2017, 33(3): 122-127. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||