








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (4): 203-216.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0685
Previous Articles Next Articles
GAO Yu-kun( ), ZHANG Jian-dong, YANG Pu-yuan, CHEN Dong-ming, WANG Zhi-bo, TIAN Yi-jin, Zakey Eldinn. E. A. Khlid, CUI Jiang-hui, CHANG Jin-hua(
), ZHANG Jian-dong, YANG Pu-yuan, CHEN Dong-ming, WANG Zhi-bo, TIAN Yi-jin, Zakey Eldinn. E. A. Khlid, CUI Jiang-hui, CHANG Jin-hua( )
)
Received:2023-07-17
Online:2024-04-26
Published:2024-04-30
Contact:
CHANG Jin-hua
E-mail:gyk1517@126.com;jhchang2006@126.com
GAO Yu-kun, ZHANG Jian-dong, YANG Pu-yuan, CHEN Dong-ming, WANG Zhi-bo, TIAN Yi-jin, Zakey Eldinn. E. A. Khlid, CUI Jiang-hui, CHANG Jin-hua. Responses of Sorghum Rhizosphere Soil Bacterial Communities to Salt Stress[J]. Biotechnology Bulletin, 2024, 40(4): 203-216.
| 指标Items | 时期Period | S0 | S3 | S7 |
|---|---|---|---|---|
| 株高 PH/cm | 拔节期 E | 67.9±6.9a | 75.8±3.8a | 54.6±4.6b |
| 开花期 F | 115.8±4.3b | 135.7±4.2a | 92.6±2.6c | |
| 成熟期 M | 164±4.6b | 185.3±4.1a | 136.5±3c | |
| 总酚TPC/(mg·100 g-1) | 拔节期 E | 60.92±19.04b | 243.92±6.03a | 209.38±23.76a |
| 开花期 F | 222.89±2.72c | 315.09±5.19a | 293.09±15.88b | |
| 成熟期 M | 240.44±53.08a | 285.43±7.59a | 250.61±7.81a | |
| 总黄酮TFC/(mg·100 g-1) | 拔节期 E | 50.5±17.02b | 180.46±20.69a | 175.15±13.72a |
| 开花期 F | 182.05±3.18c | 342.78±62.02b | 423.93±22.05a | |
| 成熟期 M | 208.04±8.17b | 273.82±11.18a | 220.24±18.35b |
Table 1 Effect of salt stress on plant height, TPC and TFC of sorghum roots
| 指标Items | 时期Period | S0 | S3 | S7 |
|---|---|---|---|---|
| 株高 PH/cm | 拔节期 E | 67.9±6.9a | 75.8±3.8a | 54.6±4.6b |
| 开花期 F | 115.8±4.3b | 135.7±4.2a | 92.6±2.6c | |
| 成熟期 M | 164±4.6b | 185.3±4.1a | 136.5±3c | |
| 总酚TPC/(mg·100 g-1) | 拔节期 E | 60.92±19.04b | 243.92±6.03a | 209.38±23.76a |
| 开花期 F | 222.89±2.72c | 315.09±5.19a | 293.09±15.88b | |
| 成熟期 M | 240.44±53.08a | 285.43±7.59a | 250.61±7.81a | |
| 总黄酮TFC/(mg·100 g-1) | 拔节期 E | 50.5±17.02b | 180.46±20.69a | 175.15±13.72a |
| 开花期 F | 182.05±3.18c | 342.78±62.02b | 423.93±22.05a | |
| 成熟期 M | 208.04±8.17b | 273.82±11.18a | 220.24±18.35b |
| 样本 Sample | 有效序列数目 Seq_num | 碱基数 Base_num/bp | 样本序列平均长度 Mean_length/nt | 样本最短序列长度 Min_length/nt | 样本最长序列长度 Max_length/nt |
|---|---|---|---|---|---|
| ES0 | 162 093 | 67 727 026 | 417.82 | 252.00 | 506.33 |
| ES3 | 160 893 | 67 266 875 | 418.08 | 244.33 | 491.67 |
| ES7 | 172 634 | 72 184 160 | 418.14 | 247.67 | 486.33 |
| FS0 | 171 403 | 71 496 233 | 417.13 | 214.33 | 468.33 |
| FS3 | 157 105 | 65 548 212 | 417.20 | 271.00 | 474.00 |
| FS7 | 161 222 | 67 470 566 | 418.48 | 235.00 | 490.67 |
| MS0 | 151 139 | 63 252 701 | 418.48 | 227.00 | 465.00 |
| MS3 | 148 582 | 61 959 724 | 417.02 | 247.33 | 477.00 |
| MS7 | 146 276 | 61 086 964 | 417.61 | 245.33 | 471.00 |
Table 2 Sequencing quality of rhizosphere microbial community
| 样本 Sample | 有效序列数目 Seq_num | 碱基数 Base_num/bp | 样本序列平均长度 Mean_length/nt | 样本最短序列长度 Min_length/nt | 样本最长序列长度 Max_length/nt |
|---|---|---|---|---|---|
| ES0 | 162 093 | 67 727 026 | 417.82 | 252.00 | 506.33 |
| ES3 | 160 893 | 67 266 875 | 418.08 | 244.33 | 491.67 |
| ES7 | 172 634 | 72 184 160 | 418.14 | 247.67 | 486.33 |
| FS0 | 171 403 | 71 496 233 | 417.13 | 214.33 | 468.33 |
| FS3 | 157 105 | 65 548 212 | 417.20 | 271.00 | 474.00 |
| FS7 | 161 222 | 67 470 566 | 418.48 | 235.00 | 490.67 |
| MS0 | 151 139 | 63 252 701 | 418.48 | 227.00 | 465.00 |
| MS3 | 148 582 | 61 959 724 | 417.02 | 247.33 | 477.00 |
| MS7 | 146 276 | 61 086 964 | 417.61 | 245.33 | 471.00 |
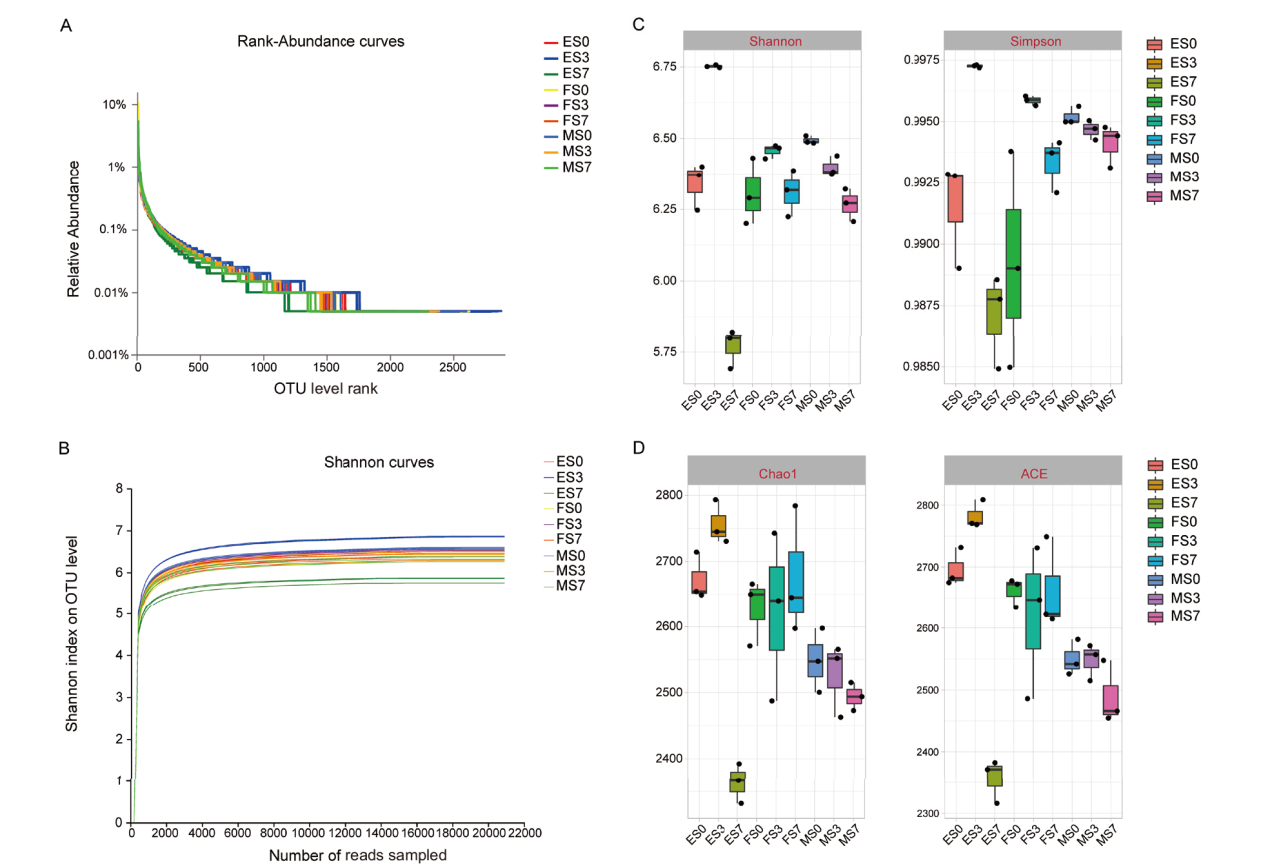
Fig. 1 Evaluation of sequencing results and Alpha diversity analysis of soil bacterial in sorghums soil samples under different treatments A: Rank abundance curve; B: dilution curves; C and D Shannon, Simpson, ACE and Chao1 indices, respectively
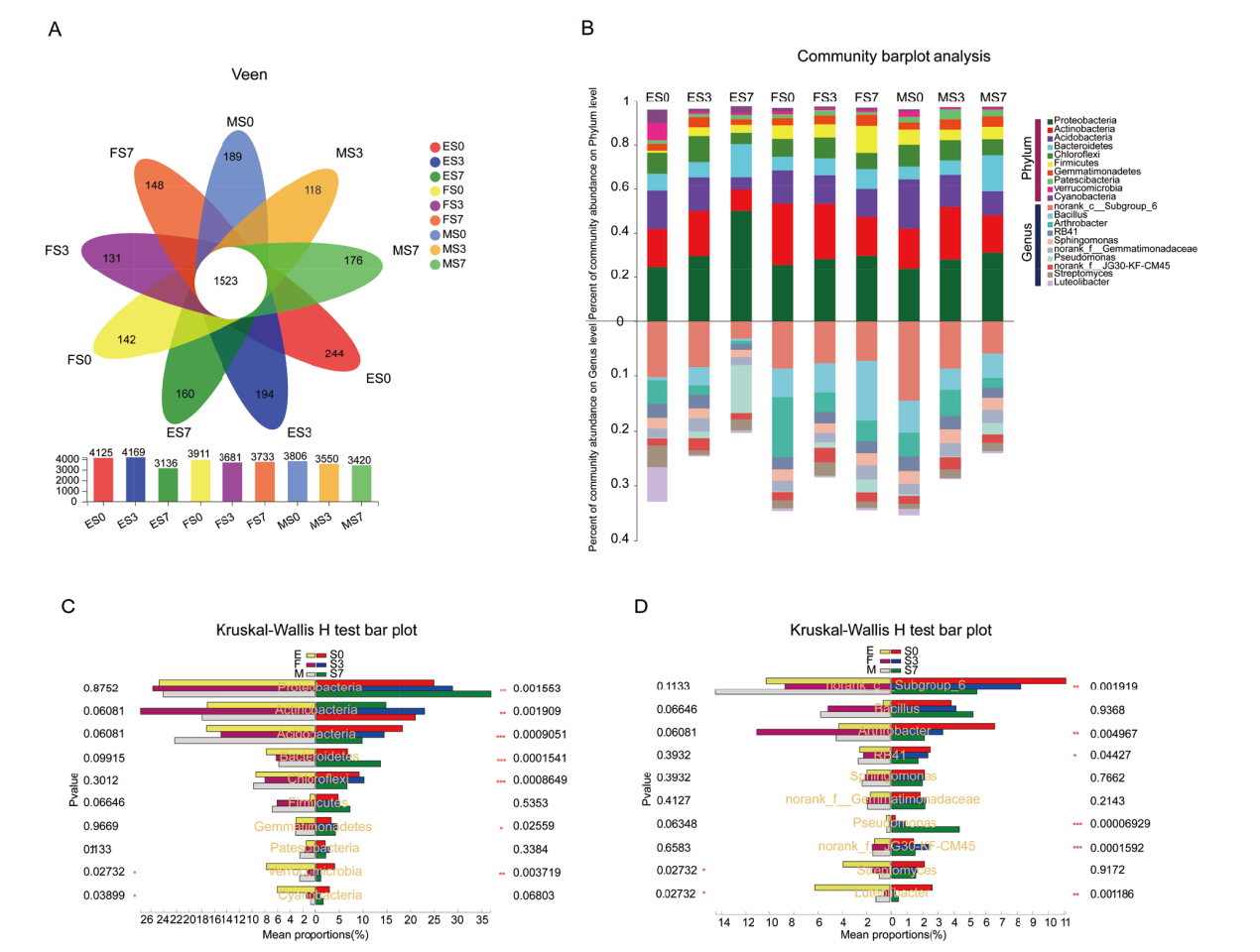
Fig. 3 Bacterial community structure of sorghum rhizosphere A: Flower plot analysis for bacterial species(OTU)of different samples. B: Map of bacterial community structure at the phylum and genus level. C and D indicate phylum and genus distribution among different groups of rhizosphere soils, respectively. *, **, and *** indicate significant differences at the 0.05, 0.01, and 0.001 levels, respectively
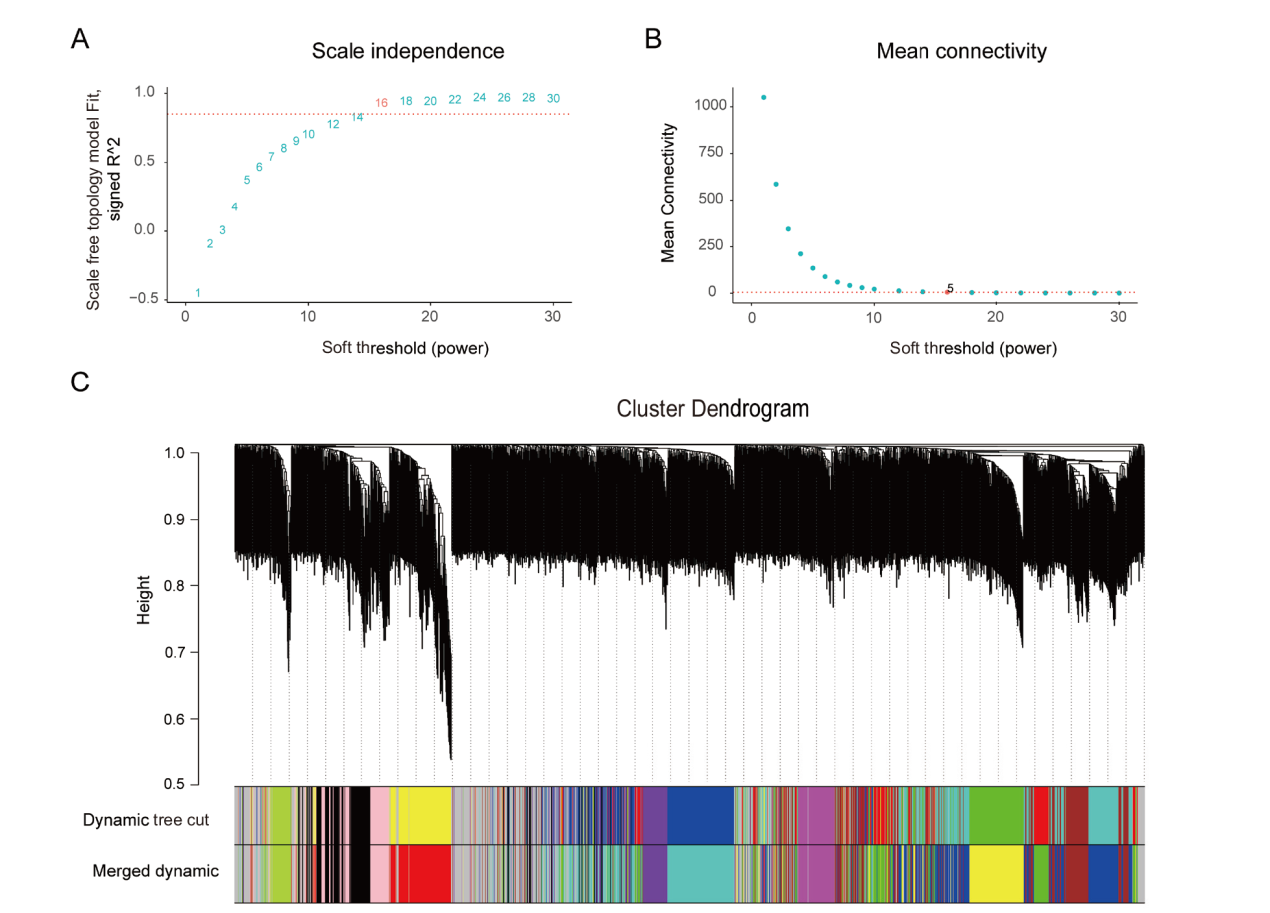
Fig. 4 Determination of soft threshold and module division The ordinate of A indicate the scale-free network model index; the ordinate of B indicate the average network connectivity corresponding to each soft threshold; the abscissas of A and B both indicate the soft threshold ; C:OTU modules obtained by dynamic shearing algorithm and after merging similar expression patterns
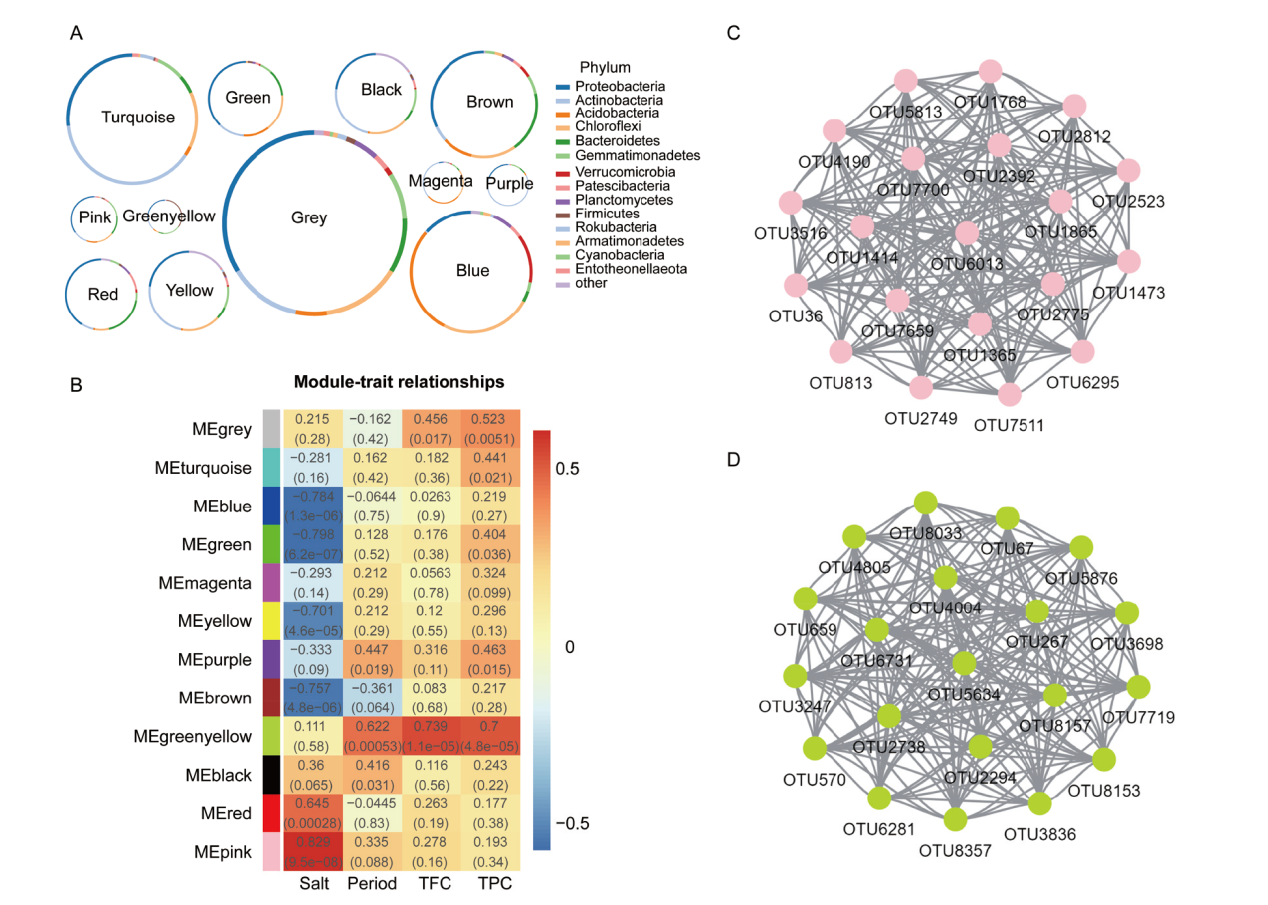
Fig. 5 Analysis of WGCNA in sorghum rhizosphere bacteria A: Bacterial phylum composition in each module. B: Association analysis of OTUs co-expression network modules with traits. C and D indicate the hub OTUs co-expression network in pink and greenyellow module, respectively
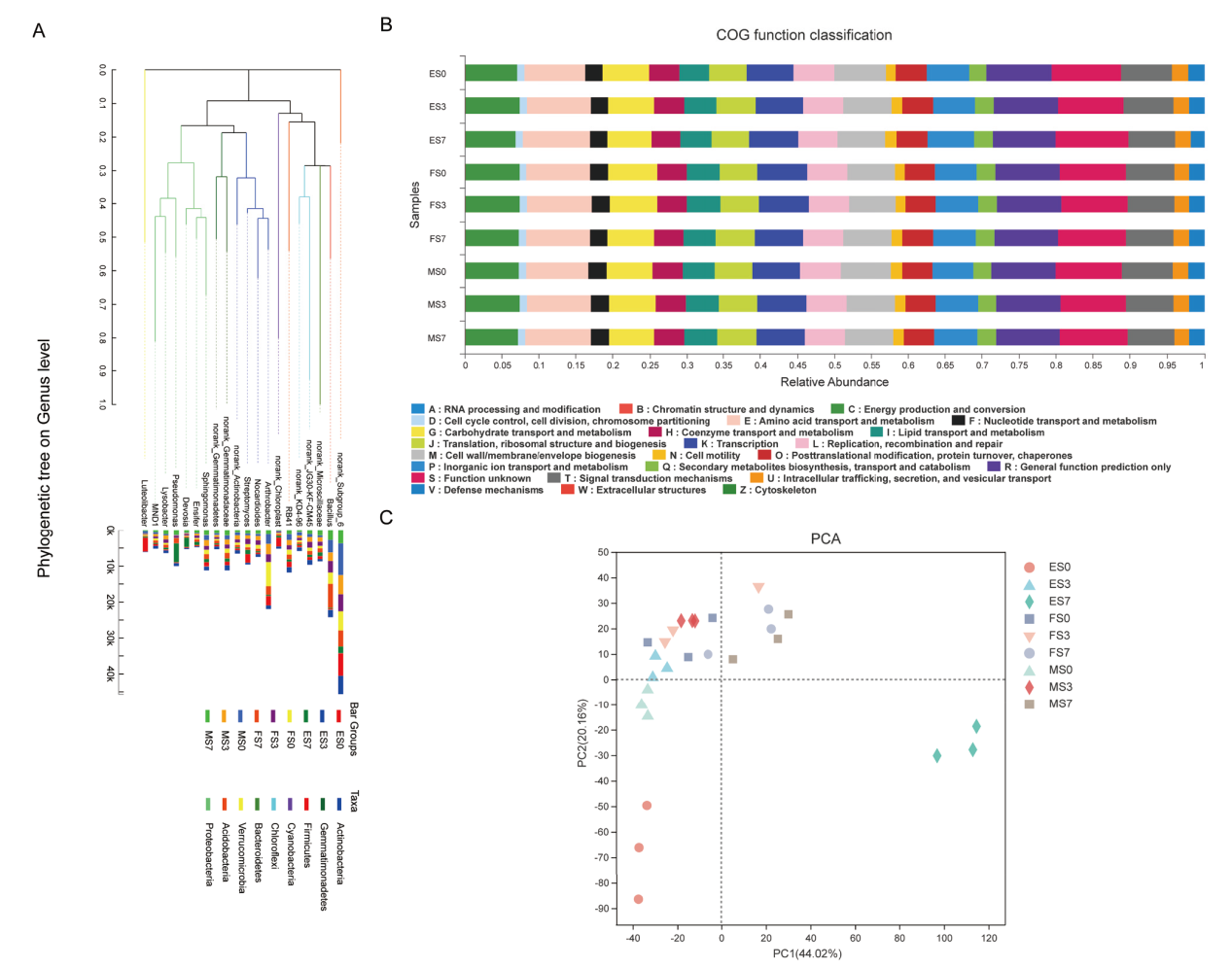
Fig. 6 A phylogenetic tree showing the relationship among salt-treated groups and microbial functional features in salt-treated soil groups via COG analysis A: Evolutionary relationship in rhizosphere soil in four salt-treated soil groups. B: The microbial functional features in three salt-treated soil groups sorghum COG analysis. C: Microbial functional features in salt-treated soil groups via COG analysis based on PCA
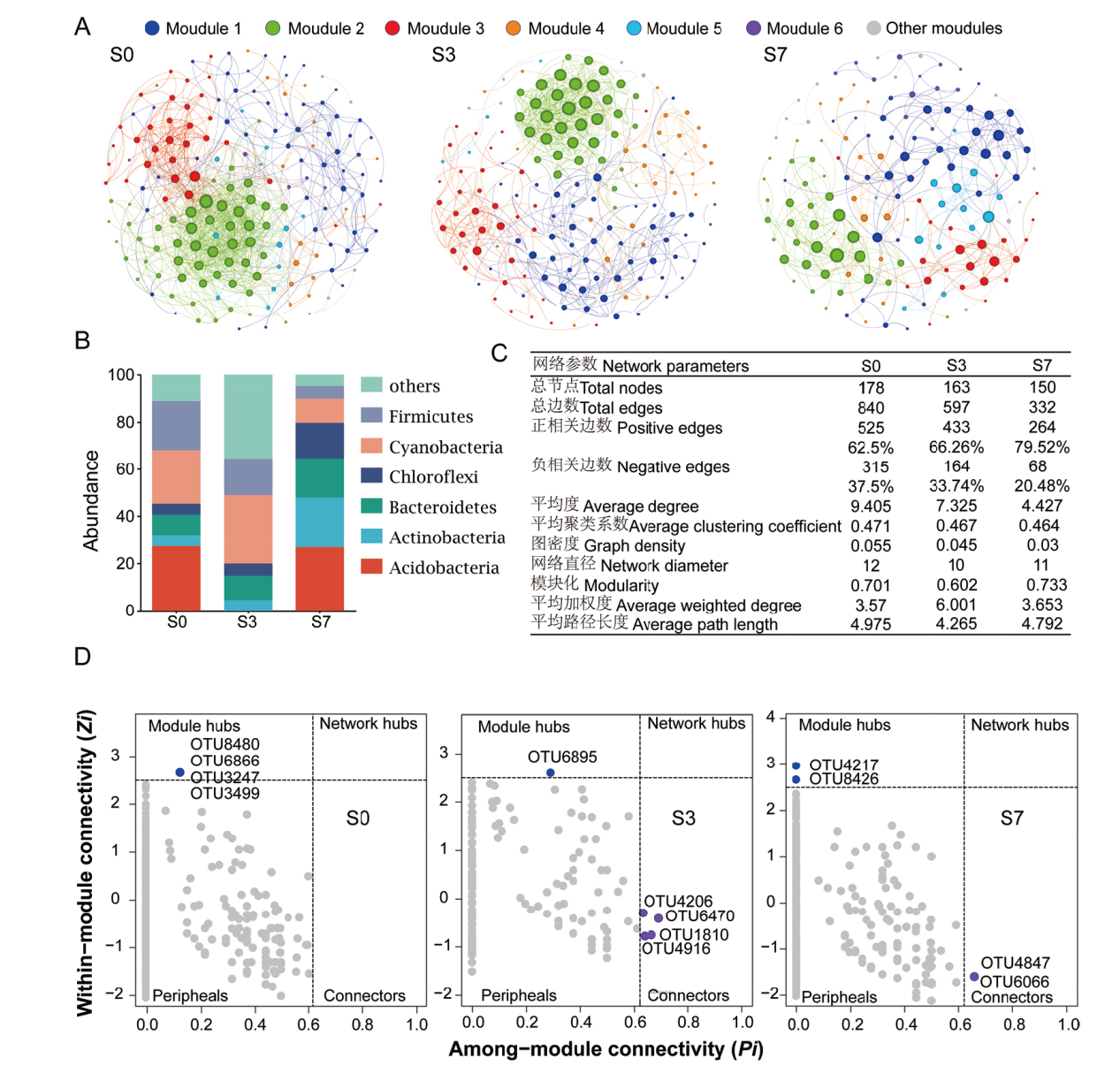
Fig. 7 Co-occurrence network in sorghum rhizosphere bacteria A: Network revealing the modular associations among sorghum rhizosphere bacterial Otus. B: Species composition of sorghum rhizosphere bacterial network under different salt stress(phylum level). C: Sorghum rhizosphere bacterial network topological characteristics under different salt stress treatments. D: Distribution of nodes based on their topological roles in modules under different salt stress treatments
| [1] |
Munns R, Tester M. Mechanisms of salinity tolerance[J]. Annu Rev Plant Biol, 2008, 59: 651-681.
doi: 10.1146/annurev.arplant.59.032607.092911 pmid: 18444910 |
| [2] |
Garthwaite AJ, von Bothmer R, Colmer TD. Salt tolerance in wild Hordeum species is associated with restricted entry of Na+ and Cl-into the shoots[J]. J Exp Bot, 2005, 56(419): 2365-2378.
pmid: 16014366 |
| [3] |
Kumar A, Verma JP. Does plant-Microbe interaction confer stress tolerance in plants: a review?[J]. Microbiol Res, 2018, 207: 41-52.
doi: S0944-5013(17)30795-4 pmid: 29458867 |
| [4] |
Zhang SY, Fan C, Wang YX, et al. Salt-tolerant and plant-growth-promoting bacteria isolated from high-yield paddy soil[J]. Can J Microbiol, 2018, 64(12): 968-978.
doi: 10.1139/cjm-2017-0571 pmid: 30148967 |
| [5] |
Berendsen RL, Pieterse CMJ, Bakker PAHM. The rhizosphere microbiome and plant health[J]. Trends Plant Sci, 2012, 17(8): 478-486.
doi: 10.1016/j.tplants.2012.04.001 pmid: 22564542 |
| [6] |
Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria[J]. Annu Rev Microbiol, 2009, 63: 541-556.
doi: 10.1146/annurev.micro.62.081307.162918 pmid: 19575558 |
| [7] |
Gouda S, Kerry RG, Das G, et al. Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture[J]. Microbiol Res, 2018, 206: 131-140.
doi: S0944-5013(17)30341-5 pmid: 29146250 |
| [8] |
戴良香, 徐扬, 张冠初, 等. 花生根际土壤细菌群落多样性对盐胁迫的响应[J]. 作物学报, 2021, 47(8): 1581-1592.
doi: 10.3724/SP.J.1006.2021.04160 |
| Dai LX, Xu Y, Zhang GC, et al. Response of rhizosphere bacterial community diversity to salt stress in peanut[J]. Acta Agron Sin, 2021, 47(8): 1581-1592. | |
| [9] |
Ahmad Ansari F, Ahmad I, Pichtel J. Growth stimulation and alleviation of salinity stress to wheat by the biofilm forming Bacillus pumilus strain FAB10[J]. Appl Soil Ecol, 2019, 143: 45-54.
doi: 10.1016/j.apsoil.2019.05.023 |
| [10] |
Tiwari S, Lata CR, Chauhan PS, et al. Pseudomonas putida attunes morphophysiological, biochemical and molecular responses in Cicer arietinum L. during drought stress and recovery[J]. Plant Physiol Biochem, 2016, 99: 108-117.
doi: 10.1016/j.plaphy.2015.11.001 URL |
| [11] |
Abdelaziz ME, Abdelsattar M, Abdeldaym EA, et al. Piriformospora indica alters Na+/K+ homeostasis, antioxidant enzymes and LeNHX1 expression of greenhouse tomato grown under salt stress[J]. Sci Hortic, 2019, 256: 108532.
doi: 10.1016/j.scienta.2019.05.059 URL |
| [12] |
Haichar FZ, Santaella C, Heulin T, et al. Root exudates mediated interactions belowground[J]. Soil Biol Biochem, 2014, 77: 69-80.
doi: 10.1016/j.soilbio.2014.06.017 URL |
| [13] |
Weir TL, Park SW, Vivanco JM. Biochemical and physiological mechanisms mediated by allelochemicals[J]. Curr Opin Plant Biol, 2004, 7(4): 472-479.
doi: 10.1016/j.pbi.2004.05.007 pmid: 15231272 |
| [14] | Minh LT, Khang DT, Thu Ha PT, et al. Effects of salinity stress on growth and phenolics of rice(Oryza sativa L.)[J]. Int Lett Nat Sci, 2016, 57: 1-10. |
| [15] | Kaur H, Bhardwaj RD, Grewal SK. Mitigation of salinity-induced oxidative damage in wheat(Triticum aestivum L.) seedlings by exogenous application of phenolic acids[J]. Acta Physiol Plant, 2017, 39(10): 221. |
| [16] | Sailaja K, Sujatha B. Impact of salt stress(nacl)on pigments, phenols and flavonoids in C4(Sorghum bicolor)and C3(oryza sativa)cultivars[J]. Int Biolo Pharm Res, 2013, 4(5): 361-367. |
| [17] |
Shaw LJ, Morris P, Hooker JE. Perception and modification of plant flavonoid signals by rhizosphere microorganisms[J]. Environ Microbiol, 2006, 8(11): 1867-1880.
pmid: 17014487 |
| [18] | 任根增, 高志远, 张瑜, 等. 混合盐碱胁迫对高粱农艺性状及生理指标的影响[J]. 作物杂志, 2017(1): 100-106. |
| Ren GZ, Gao ZY, Zhang Y, et al. The influence of mixed salt and alkali stress on physiological and agronomic traits of Sorghum[J]. Crops, 2017(1): 100-106. | |
| [19] |
高玉坤, 杨溥原, 项晓冬, 等. 不同耐盐高粱品种全生育期对盐胁迫的响应[J]. 华北农学报, 2020, 35(6): 113-121.
doi: 10.7668/hbnxb.20191411 |
| Gao YK, Yang PY, Xiang XD, et al. Response of different salt tolerant sorghum varieties to salt stress in the whole growth period[J]. Acta Agric Boreali Sin, 2020, 35(6): 113-121. | |
| [20] |
Cui JH, Ren GZ, Qiao HY, et al. Comparative transcriptome analysis of seedling stage of two sorghum cultivars under salt stress[J]. J Plant Growth Regul, 2018, 37(3): 986-998.
doi: 10.1007/s00344-018-9796-9 |
| [21] |
Huang R, Zhang Y, Shen SY, et al. Antioxidant and pancreatic lipase inhibitory effects of flavonoids from different citrus peel extracts: an in vitro study[J]. Food Chem, 2020, 326: 126785.
doi: 10.1016/j.foodchem.2020.126785 URL |
| [22] |
Kaur N, Singh B, Kaur A, et al. Effect of growing conditions on proximate, mineral, amino acid, phenolic composition and antioxidant properties of wheatgrass from different wheat(Triticum aestivum L.) varieties[J]. Food Chem, 2021, 341(Pt 1): 128201.
doi: 10.1016/j.foodchem.2020.128201 URL |
| [23] | Gao YK, Cui JH, Ren GZ, et al. Changes in the root-associated bacteria of sorghum are driven by the combined effects of salt and sorghum development[J]. Environ Microbiome, 2021, 16(1): 14. |
| [24] |
Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data[J]. Nat Methods, 2010, 7(5): 335-336.
doi: 10.1038/nmeth.f.303 pmid: 20383131 |
| [25] |
Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads[J]. Nat Methods, 2013, 10(10): 996-998.
doi: 10.1038/nmeth.2604 pmid: 23955772 |
| [26] | Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools[J]. Nucleic Acids Res, 2013, 41(Database issue): D590-D596. |
| [27] | Weiss S, Xu ZZ, Peddada S, et al. Normalization and microbial differential abundance strategies depend upon data characteristics[J]. Microbiome, 2017, 5(1): 27. |
| [28] |
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis[J]. BMC Bioinformatics, 2008, 9: 559.
doi: 10.1186/1471-2105-9-559 pmid: 19114008 |
| [29] |
Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks[J]. Genome Res, 2003, 13(11): 2498-2504.
doi: 10.1101/gr.1239303 pmid: 14597658 |
| [30] |
Bai Y, Müller DB, Srinivas G, et al. Functional overlap of the Ara-bidopsis leaf and root microbiota[J]. Nature, 2015, 528(7582): 364-369.
doi: 10.1038/nature16192 |
| [31] | Csárdi G, Nepusz T. The igraph software package for complex network research[J]. Inter Journal Complex Systems, 2006, 1695: 1-9. |
| [32] |
Deng Y, Jiang YH, Yang YF, et al. Molecular ecological network analyses[J]. BMC Bioinformatics, 2012, 13: 113.
doi: 10.1186/1471-2105-13-113 pmid: 22646978 |
| [33] |
Sarker U, Oba S. Salinity stress enhances color parameters, bioactive leaf pigments, vitamins, polyphenols, flavonoids and antioxidant activity in selected Amaranthus leafy vegetables[J]. J Sci Food Agric, 2019, 99(5): 2275-2284.
doi: 10.1002/jsfa.2019.99.issue-5 URL |
| [34] |
Yang YQ, Guo Y. Unraveling salt stress signaling in plants[J]. J Integr Plant Biol, 2018, 60(9): 796-804.
doi: 10.1111/jipb.12689 |
| [35] |
Zhang JL, Shi HZ. Physiological and molecular mechanisms of plant salt tolerance[J]. Photosynth Res, 2013, 115(1): 1-22.
doi: 10.1007/s11120-013-9813-6 URL |
| [36] | 宁亚茹, 晋梦珂, 王秀萍, 等. 盐胁迫对黄蜀葵生长生理指标及总黄酮含量的影响[J]. 中药材, 2020, 43(2): 259-263. |
| Ning YR, Jin MK, Wang XP, et al. Effects of salt stress on growth physiological indicators and total flavonoids content of Abelmoschus manihot[J]. J Chin Med Mater, 2020, 43(2): 259-263. | |
| [37] |
Bistgani ZE, Hashemi M, DaCosta M, et al. Effect of salinity stress on the physiological characteristics, phenolic compounds and antioxidant activity of Thymus vulgaris L. and Thymus daenensis Celak[J]. Ind Crops Prod, 2019, 135: 311-320.
doi: 10.1016/j.indcrop.2019.04.055 URL |
| [38] |
Salem N, Msaada K, Dhifi W, et al. Effect of salinity on plant growth and biological activities of Carthamus tinctorius L. extracts at two flowering stages[J]. Acta Physiol Plant, 2014, 36(2): 433-445.
doi: 10.1007/s11738-013-1424-5 URL |
| [39] |
Farhadi N, Babaei K, Farsaraei S, et al. Changes in essential oil compositions, total phenol, flavonoids and antioxidant capacity of Achillea millefolium at different growth stages[J]. Ind Crops Prod, 2020, 152: 112570.
doi: 10.1016/j.indcrop.2020.112570 URL |
| [40] |
Brimecombe MJ, Leij FA, Lynch JM. Nematode community structure as a sensitive indicator of microbial perturbations induced by a genetically modified Pseudomonas fluorescens strain[J]. Biol Fertil Soils, 2001, 34(4): 270-275.
doi: 10.1007/s003740100412 URL |
| [41] |
Lucas García JA, Barbas C, Probanza A, et al. Low molecular weight organic acids and fatty acids in root exudates of two Lupinus cultivars at flowering and fruiting stages[J]. Phytochem Anal, 2001, 12(5): 305-311.
doi: 10.1002/pca.v12:5 URL |
| [42] | Xu L, Naylor D, Dong ZB, et al. Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria[J]. Proc Natl Acad Sci USA, 2018, 115(18): E4284-E4293. |
| [43] |
Bulgarelli D, Garrido-Oter R, Münch PC, et al. Structure and function of the bacterial root microbiota in wild and domesticated barley[J]. Cell Host Microbe, 2015, 17(3): 392-403.
doi: S1931-3128(15)00031-1 pmid: 25732064 |
| [44] |
Lundberg DS, Lebeis SL, Paredes SH, et al. Defining the core Arabidopsis thaliana root microbiome[J]. Nature, 2012, 488(7409): 86-90.
doi: 10.1038/nature11237 |
| [45] |
郭雨晴, 赵世超, 徐道龙, 等. 3种荒漠珍稀植物根际促生菌的筛选、鉴定及对高粱幼苗生长的影响[J]. 草地学报, 2020, 28(4): 1121-1128.
doi: 10.11733/j.issn.1007-0435.2020.04.031 |
| Guo YQ, Zhao SC, Xu DL, et al. Screening and identification of growth-promoting bacteria from three rare plants rhizosphere soil in desert and its effect on the growth of sorghum seedlings[J]. Acta Agrestia Sin, 2020, 28(4): 1121-1128. | |
| [46] |
Zelm E, Zhang YX, Testerink C. Salt tolerance mechanisms of plants[J]. Annu Rev Plant Biol, 2020, 71: 403-433.
doi: 10.1146/annurev-arplant-050718-100005 pmid: 32167791 |
| [47] |
Nautiyal CS, Srivastava S, Chauhan PS, et al. Plant growth-promoting bacteria Bacillus amyloliquefaciens NBRISN13 modulates gene expression profile of leaf and rhizosphere community in rice during salt stress[J]. Plant Physiol Biochem, 2013, 66: 1-9.
doi: 10.1016/j.plaphy.2013.01.020 URL |
| [48] |
Chen L, Liu YP, Wu GW, et al. Induced maize salt tolerance by rhizosphere inoculation of Bacillus amyloliquefaciens SQR9[J]. Physiol Plant, 2016, 158(1): 34-44.
doi: 10.1111/ppl.12441 pmid: 26932244 |
| [49] |
Wang WF, Wu ZS, He YH, et al. Plant growth promotion and alleviation of salinity stress in Capsicum annuum L. by Bacillus isolated from saline soil in Xinjiang[J]. Ecotoxicol Environ Saf, 2018, 164: 520-529.
doi: 10.1016/j.ecoenv.2018.08.070 URL |
| [50] |
Yan JM, Smith MD, Glick BR, et al. Effects of ACC deaminase containing rhizobacteria on plant growth and expression of Toc GTPases in tomato(Solanum lycopersicum)under salt stress[J]. Botany, 2014, 92(11): 775-781.
doi: 10.1139/cjb-2014-0038 URL |
| [51] | Rahman M, Sabir AA, Mukta JA, et al. Plant probiotic bacteria Bacillus and Paraburkholderia improve growth, yield and content of antioxidants in strawberry fruit[J]. Sci Rep, 2018, 8(1): 2504. |
| [52] | Chandrasekaran M, Chun SC, Oh JW, et al. Bacillus subtilis CBR05 for tomato(Solanum lycopersicum)fruits in South Korea as a novel plant probiotic bacterium(PPB): implications from total phenolics, flavonoids, and carotenoids content for fruit quality[J]. Agronomy, 2019, 9(12): 838. |
| [53] |
An XC, Wang ZF, Teng XM, et al. Rhizosphere bacterial diversity and environmental function prediction of wild salt-tolerant plants in coastal silt soil[J]. Ecol Indic, 2022, 134: 108503.
doi: 10.1016/j.ecolind.2021.108503 URL |
| [54] | Paranjape K, Bédard É, Shetty D, et al. Unravelling the importance of the eukaryotic and bacterial communities and their relationship with Legionella spp. ecology in cooling towers: a complex network[J]. Microbiome, 2020, 8(1): 157. |
| [55] |
Lu LH, Yin SX, Liu X, et al. Fungal networks in yield-invigorating and-debilitating soils induced by prolonged potato monoculture[J]. Soil Biol Biochem, 2013, 65: 186-194.
doi: 10.1016/j.soilbio.2013.05.025 URL |
| [56] |
Shi SJ, Nuccio EE, Shi ZJ, et al. The interconnected rhizosphere: high network complexity dominates rhizosphere assemblages[J]. Ecol Lett, 2016, 19(8): 926-936.
doi: 10.1111/ele.12630 pmid: 27264635 |
| [1] | QI Yan-xiang, XIE Yi-xian, PENG Jun, ZENG Fan-yun, ZHANG Xin. Banana Rhizosphere Microecology and Its Relationship with Fusarium Wilt Control [J]. Biotechnology Bulletin, 2024, 40(6): 57-67. |
| [2] | LI Jing-yan, ZHOU Jia-jing, YUAN Yuan, SU Xiao-yi, QIAO Wen-hui, XUE Yan-lei, LI Guo-jing, WANG Rui-gang. Response of Arabidopsis AtiPGAM2 Gene to Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(5): 215-224. |
| [3] | YU Li-jun, WANG Qiao-mei, PENG Wen-shu, YAN Liang, YANG Rui-juan. Study on the Microbial Community of Rhizosphere Soil in Ancient Tea Garden and Modern Organic Tea Garden in Jingmai Mountain [J]. Biotechnology Bulletin, 2024, 40(5): 237-247. |
| [4] | LI Hui, WEN Yu-fang, WANG Yue, JI Chao, SHI Guo-you, LUO Ying, ZHOU Yong, LI Zhi-min, WU Xiao-yu, YANG You-xin, LIU Jian-ping. Expression Characteristics and Functions of CaPIF4 in Capsicum annuum Under Salt Stress [J]. Biotechnology Bulletin, 2024, 40(4): 148-158. |
| [5] | GAO Zhi-wei, WEI Ming, YU Zu-long, WU Guo-qiang, WEI Jun-long. Identification of Salt-tolerant Plant Growth-promoting Bacterium W-1 and Its Effect on the Salt-tolerance of Sainfoin(Onobrychis viciaefolia) [J]. Biotechnology Bulletin, 2024, 40(4): 217-227. |
| [6] | WANG Hao-jie, CHANG Dong, LI Jun-ying, MENG Hao-guang, JIANG Shi-jun, ZHOU Shuo-ye, CUI Jiang-kuan. Analysis of Microbial Community Changes and Stress-resistant Enzyme Activities of Flue-cured Tobacco Three-stage Seedling Raising in Different Habitats [J]. Biotechnology Bulletin, 2024, 40(4): 242-254. |
| [7] | LIU Jia-ning, LI Meng, YANG Xin-sen, WU Wei, PEI Xin-wu, YUAN Qian-hua. Impact of Different Water Management Cultivation Methods on the Rhizosphere Bacteria Community of Shanlan Upland Rice [J]. Biotechnology Bulletin, 2024, 40(3): 242-250. |
| [8] | SHEN Tian-hong, QI Xiao-bo, ZHAO Rui-feng, MA Xin-rong. Research Progress in the Molecular Mechanisms of Microalgae Responding to Salt Stress [J]. Biotechnology Bulletin, 2024, 40(3): 89-99. |
| [9] | LI Xue, LI Rong-ou, KONG Mei-yi, HUANG Lei. The Growth Promoting Effect of Bacillus amyloliquefaciens SQ-2 on Rice [J]. Biotechnology Bulletin, 2024, 40(2): 109-119. |
| [10] | LI Hao, WU Guo-qiang, WEI Ming, HAN Yue-xin. Genome-wide Identification of the BvBADH Gene Family in Sugar Beet(Beta vulgaris)and Their Expression Analysis Under High Salt Stress [J]. Biotechnology Bulletin, 2024, 40(2): 233-244. |
| [11] | XU Yang, ZHANG Rui-ying, DAI Liang-xiang, ZHANG Guan-chu, DING Hong, ZHANG Zhi-meng. Regulation of Nitrogen Application on Peanut Seed Germination and Spermosphere Bacterial Community Structure Under Salt Stress [J]. Biotechnology Bulletin, 2024, 40(2): 253-265. |
| [12] | LEI Mei-ling, RAO Wen-hua, HU Jin-feng, YUE Qi, WU Zu-jian, FAN Guo-cheng. Bacterial Diversity and Structure in Rhizosphere Soil of Citrus Infested with Huanglongbing [J]. Biotechnology Bulletin, 2024, 40(2): 266-276. |
| [13] | WANG Yu-qing, MA Zi-qi, HOU Jia-xin, ZONG Yu-qi, HAO Han-rui, LIU Guo-yuan, WEI Hui, LIAN Bo-lin, CHEN Yan-hong, ZHANG Jian. Research Progress in the Composition Analysis and Ecological Function of Plant Root Exudates Under Salt Stress [J]. Biotechnology Bulletin, 2024, 40(1): 12-23. |
| [14] | FENG Lu-yao, ZHAO Jiang-yuan, SHI Zhu-feng, MO Yan-fang, YANG Tong-yu, SHEN Yun-xin, HE Fei-fei, LI Ming-gang, YANG Pei-wen. Isolation and Identification of Bacteria in Forest Rhizosphere Soil and Their Biological Activity Screening [J]. Biotechnology Bulletin, 2024, 40(1): 294-307. |
| [15] | ZHAO Guang-xu, YANG He-tong, SHAO Xiao-bo, CUI Zhi-hao, LIU Hong-guang, ZHANG Jie. Phosphate-solubilizing Properties and Optimization of Cultivation Conditions of Penicillium rubens: A Highly Efficient Phosphate Solubilizer [J]. Biotechnology Bulletin, 2023, 39(9): 71-83. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||