








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (6): 114-125.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1184
Previous Articles Next Articles
ZHANG Di1( ), JU Rui1, LI Li-mei2, WANG Yu-qian3, CHEN Rui3, WANG Xin-yi1(
), JU Rui1, LI Li-mei2, WANG Yu-qian3, CHEN Rui3, WANG Xin-yi1( )
)
Received:2023-12-15
Online:2024-06-26
Published:2024-04-19
Contact:
WANG Xin-yi
E-mail:deezhang163@163.com;wangxinyi@dlu.edu.cn
ZHANG Di, JU Rui, LI Li-mei, WANG Yu-qian, CHEN Rui, WANG Xin-yi. Application of Transcription Factor-based Biosensors in Environmental Analysis[J]. Biotechnology Bulletin, 2024, 40(6): 114-125.
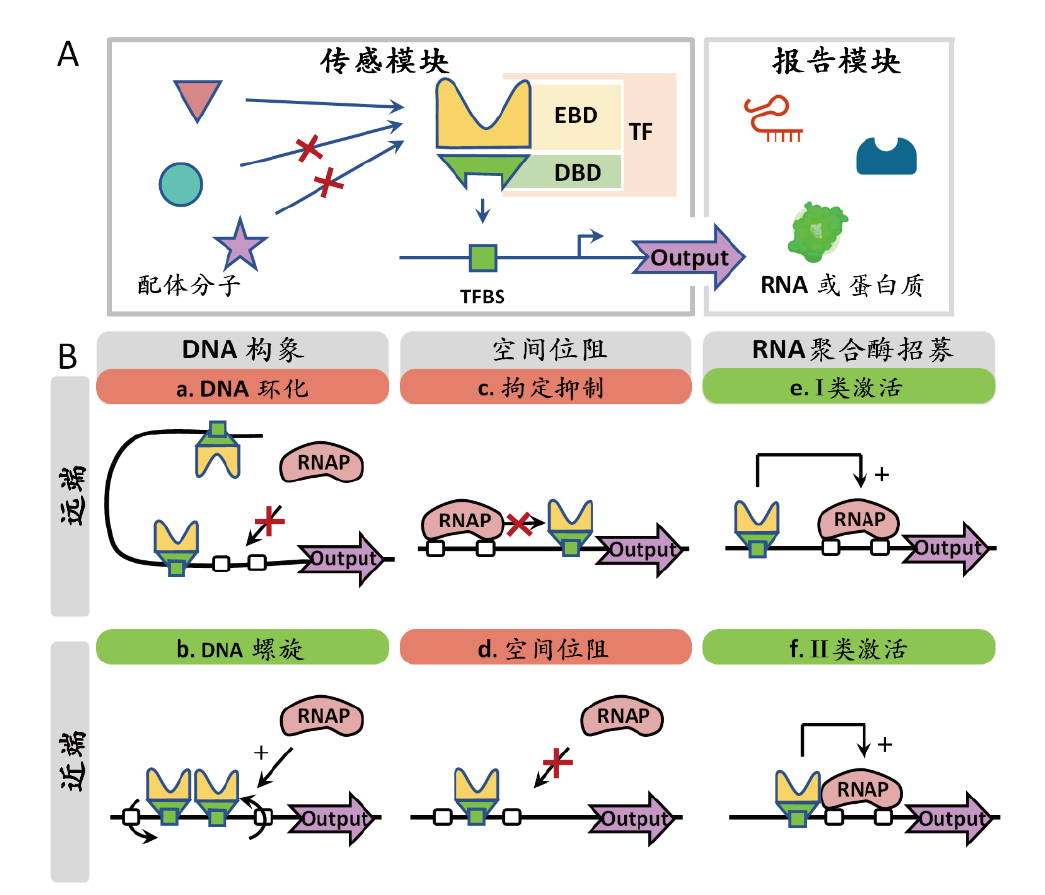
Fig. 1 Principle of transcription factor-based biosensors A: Schematic diagram of biosensor structure; B: schematic description of transcription factor activation and repression mechanisms
| 重金属 Heavy metal ion | 转录因子/家族 TFs/families | PDB | 反应平台 Reaction platform | 报告元件 Report component | 检测限 Limit of detection | 参考文献 Reference |
|---|---|---|---|---|---|---|
| 汞Hg(II) | MerR/MerR | - | 全细胞平台 | mCherry | 0.1 nmol/L | [ |
| 全细胞平台 | GFP | 4.4 nmol/L | [ | |||
| 全细胞平台 | RFP | 1 mg/kg | [ | |||
| 无细胞平台 | 3WJdB | 0.5 nmol/L | [ | |||
| 全细胞平台 | sfCherry3C | 2.24 µmol/L | [ | |||
| CadR/MerR | - | 全细胞平台 | GFP、RFP | 0.01 µg/mL | [ | |
| 镉Cd(II) | ZntR/MerR | 1Q08 | 全细胞平台 | sGFP | 35 nmol/L | [ |
| 全细胞平台 | eGFP | 1 000 nmol/L | [ | |||
| 全细胞平台 | Lux | 100 nmol/L | [ | |||
| MerR/MerR | - | 全细胞平台 | GFP | 283.9 nmol/L | [ | |
| 全细胞平台 | eGFP | 3 µmol/L | [ | |||
| CadC/ArsR/SmtB | 1U2W | 全细胞平台 | GFP | 10 µg/mL | [ | |
| CadR/MerR | - | 全细胞平台 | mCherry | 0.1 µg/mL | [ | |
| ZntR/MerR | 1Q08 | 全细胞平台 | GFP | 5 µmol/L | [ | |
| 砷As(III) | ArsR/ArsR/SmtB | 1R23 | 邻近效应驱动 | Lac Z | 8 ppb | [ |
| ArsR/ArsR/SmtB | - | 全细胞平台 | GFP | 0.1 µmol/L | [ | |
| CadR/MerR | - | 全细胞平台 | GFP、RFP | 0.01 µg/mL | [ | |
| 铜Cu(II) | CopR/CopY | - | 全细胞平台 | RFP | 24.3 µmol/L | [ |
| CueR/MerR | 1Q05 | 全细胞平台 | eGFP | 0.08 µmol/L | [ | |
| CusR | - | 全细胞平台 | GFP | 12 µmol/L | [ | |
| 锌Zn(II) | SmtB/ArsR/SmtB | 1R23 | 全细胞平台 | eGFP | 1 µmol/L | [ |
| ZntR/MerR | 1Q08 | 全细胞平台 | GFP、mCherry | 5 mg/L | [ | |
| MerRZntR/MerR | - | 全细胞平台 | eGFP、mCherry | 10 µg/mL | [ | |
| 全细胞平台 | Lux | 0.03 mmol/L | [ | |||
| 铅Pb(II) | CadR/MerR | - | 全细胞平台 | GFP、RFP | 0.01 µg/mL | [ |
| MerR/MerR | - | 全细胞平台 | GFP | 39.6 nmol/L | [ | |
| PbrR/MerR | - | 全细胞平台 | 靛蓝合成酶Indigoidine synthetase | 0.065 mol/L | [ | |
| 无细胞平台 | 3WJdB | 0.1 nmol/L | [ | |||
| 全细胞平台 | 比色法 | 2.93 nmol/L | [ | |||
| 锑Sb(III) | ArsR/ArsR/SmtB | 1R23 | 全细胞平台 | eGFP | 0.25 µmol/L | [ |
| 镍Ni(II) | RcnR/CsoR-RcnR | - | 全细胞平台 | Lux | 80 nmol/L | [ |
Table 1 Detection of heavy metal ions by transcription factor-based biosensors
| 重金属 Heavy metal ion | 转录因子/家族 TFs/families | PDB | 反应平台 Reaction platform | 报告元件 Report component | 检测限 Limit of detection | 参考文献 Reference |
|---|---|---|---|---|---|---|
| 汞Hg(II) | MerR/MerR | - | 全细胞平台 | mCherry | 0.1 nmol/L | [ |
| 全细胞平台 | GFP | 4.4 nmol/L | [ | |||
| 全细胞平台 | RFP | 1 mg/kg | [ | |||
| 无细胞平台 | 3WJdB | 0.5 nmol/L | [ | |||
| 全细胞平台 | sfCherry3C | 2.24 µmol/L | [ | |||
| CadR/MerR | - | 全细胞平台 | GFP、RFP | 0.01 µg/mL | [ | |
| 镉Cd(II) | ZntR/MerR | 1Q08 | 全细胞平台 | sGFP | 35 nmol/L | [ |
| 全细胞平台 | eGFP | 1 000 nmol/L | [ | |||
| 全细胞平台 | Lux | 100 nmol/L | [ | |||
| MerR/MerR | - | 全细胞平台 | GFP | 283.9 nmol/L | [ | |
| 全细胞平台 | eGFP | 3 µmol/L | [ | |||
| CadC/ArsR/SmtB | 1U2W | 全细胞平台 | GFP | 10 µg/mL | [ | |
| CadR/MerR | - | 全细胞平台 | mCherry | 0.1 µg/mL | [ | |
| ZntR/MerR | 1Q08 | 全细胞平台 | GFP | 5 µmol/L | [ | |
| 砷As(III) | ArsR/ArsR/SmtB | 1R23 | 邻近效应驱动 | Lac Z | 8 ppb | [ |
| ArsR/ArsR/SmtB | - | 全细胞平台 | GFP | 0.1 µmol/L | [ | |
| CadR/MerR | - | 全细胞平台 | GFP、RFP | 0.01 µg/mL | [ | |
| 铜Cu(II) | CopR/CopY | - | 全细胞平台 | RFP | 24.3 µmol/L | [ |
| CueR/MerR | 1Q05 | 全细胞平台 | eGFP | 0.08 µmol/L | [ | |
| CusR | - | 全细胞平台 | GFP | 12 µmol/L | [ | |
| 锌Zn(II) | SmtB/ArsR/SmtB | 1R23 | 全细胞平台 | eGFP | 1 µmol/L | [ |
| ZntR/MerR | 1Q08 | 全细胞平台 | GFP、mCherry | 5 mg/L | [ | |
| MerRZntR/MerR | - | 全细胞平台 | eGFP、mCherry | 10 µg/mL | [ | |
| 全细胞平台 | Lux | 0.03 mmol/L | [ | |||
| 铅Pb(II) | CadR/MerR | - | 全细胞平台 | GFP、RFP | 0.01 µg/mL | [ |
| MerR/MerR | - | 全细胞平台 | GFP | 39.6 nmol/L | [ | |
| PbrR/MerR | - | 全细胞平台 | 靛蓝合成酶Indigoidine synthetase | 0.065 mol/L | [ | |
| 无细胞平台 | 3WJdB | 0.1 nmol/L | [ | |||
| 全细胞平台 | 比色法 | 2.93 nmol/L | [ | |||
| 锑Sb(III) | ArsR/ArsR/SmtB | 1R23 | 全细胞平台 | eGFP | 0.25 µmol/L | [ |
| 镍Ni(II) | RcnR/CsoR-RcnR | - | 全细胞平台 | Lux | 80 nmol/L | [ |
| 芳香族化合物 Aromatic compound | 转录因子/家族 TFs/families | PDB | 反应平台 Reaction platform | 报告元件 Report component | 检测限 Limit of detection | 参考文献 Reference |
|---|---|---|---|---|---|---|
| 苯甲酸 | BenR/XylS-AraC | - | 无细胞平台 | sfGFP | 1 µmol/L | [ |
| - | 无细胞平台 | GFP | 21 702.8 nmol/L | [ | ||
| 苯 | DmpR/XylR-NtrC | 5KBE | 体外酶放大系统 | ATP酶 | 0.3 ppm | [ |
| 氯苯酚 | DmpR/XylR-NtrC | 全细胞平台 | Lac Z | 100 µmol/L | [ | |
| 苯并芘 | DmpR/XylR-NtrC | 全细胞平台 | eGFP | 2.5 ppb | [ | |
| 甲苯 | XylR/XylR-NtrC | 4FE4 | 全细胞平台 | 荧光素酶 | 1 µmol/L | [ |
| 2,4-二硝基甲苯 | XylR/XylR-NtrC | 全细胞平台 | GFP/lux | ~µmol/L | [ | |
| XylRv17/XylR-NtrC | - | 全细胞平台 | 荧光素酶/lacZ | - | [ | |
| 2-羟基-3',4'-二氯联苯 | HbpR/XylR-NtrC | - | 全细胞平台 | 荧光素酶 | 10 nmol/L | [ |
| 苯酚 | MopR/XylR-NtrC | 5KBE | 全细胞平台 | 荧光素酶 | 1 ppb | [ |
| 4-硝基苯酚 | Dm01/Dm12/XylR-NtrC | - | 全细胞平台 | RFP | 10 µmol/L | [ |
| 水杨酸 | SAR2349/NahR-LysR | 4EM0 | 无细胞平台 | 3WJdB荧光适配体/sfGFP | ~µmol/L | [ |
| 苯并芘 | SalR/NahR-LysR | - | 全细胞平台 | 荧光素酶 | 0.01 µmol/L | [ |
| 萘 | NahR/NahR-LysR | - | 全细胞平台 | 荧光素酶 | 50 nmol/L | [ |
| 水杨酸 | CmeR/TetR | - | 全细胞平台 | eGFP | ~µmol/L | [ |
| 苯扎氯氨 | QacR/TetR/CamR | 1JT0 | 无细胞平台 | 3WJdB荧光适配体 | ~µmol/L | [ |
| 苯甲酸盐 | MarR/MarR | 1JGS | 全细胞平台 | yEGFP | ~ fmol/L | [ |
| 苯甲醛 | BldR/MarR | 3F3X | 全细胞平台 | eGFP | ~µmol/L | [ |
| 3-羟基苯甲酸 | MobR/MarR | - | 无细胞平台 | 3WJdB荧光适配体/ | - | [ |
| 苯亚胂酸盐 | ArsR/ArsR | 6J05 | 全细胞平台 | GFP | 1 µmol/L | [ |
| 丙酮酸盐 | PdhR/GntR | - | 全细胞平台 | yEGFP | ~ fmol/L | [ |
Table 2 Detection of aromatic compounds by transcription factor-based biosensors
| 芳香族化合物 Aromatic compound | 转录因子/家族 TFs/families | PDB | 反应平台 Reaction platform | 报告元件 Report component | 检测限 Limit of detection | 参考文献 Reference |
|---|---|---|---|---|---|---|
| 苯甲酸 | BenR/XylS-AraC | - | 无细胞平台 | sfGFP | 1 µmol/L | [ |
| - | 无细胞平台 | GFP | 21 702.8 nmol/L | [ | ||
| 苯 | DmpR/XylR-NtrC | 5KBE | 体外酶放大系统 | ATP酶 | 0.3 ppm | [ |
| 氯苯酚 | DmpR/XylR-NtrC | 全细胞平台 | Lac Z | 100 µmol/L | [ | |
| 苯并芘 | DmpR/XylR-NtrC | 全细胞平台 | eGFP | 2.5 ppb | [ | |
| 甲苯 | XylR/XylR-NtrC | 4FE4 | 全细胞平台 | 荧光素酶 | 1 µmol/L | [ |
| 2,4-二硝基甲苯 | XylR/XylR-NtrC | 全细胞平台 | GFP/lux | ~µmol/L | [ | |
| XylRv17/XylR-NtrC | - | 全细胞平台 | 荧光素酶/lacZ | - | [ | |
| 2-羟基-3',4'-二氯联苯 | HbpR/XylR-NtrC | - | 全细胞平台 | 荧光素酶 | 10 nmol/L | [ |
| 苯酚 | MopR/XylR-NtrC | 5KBE | 全细胞平台 | 荧光素酶 | 1 ppb | [ |
| 4-硝基苯酚 | Dm01/Dm12/XylR-NtrC | - | 全细胞平台 | RFP | 10 µmol/L | [ |
| 水杨酸 | SAR2349/NahR-LysR | 4EM0 | 无细胞平台 | 3WJdB荧光适配体/sfGFP | ~µmol/L | [ |
| 苯并芘 | SalR/NahR-LysR | - | 全细胞平台 | 荧光素酶 | 0.01 µmol/L | [ |
| 萘 | NahR/NahR-LysR | - | 全细胞平台 | 荧光素酶 | 50 nmol/L | [ |
| 水杨酸 | CmeR/TetR | - | 全细胞平台 | eGFP | ~µmol/L | [ |
| 苯扎氯氨 | QacR/TetR/CamR | 1JT0 | 无细胞平台 | 3WJdB荧光适配体 | ~µmol/L | [ |
| 苯甲酸盐 | MarR/MarR | 1JGS | 全细胞平台 | yEGFP | ~ fmol/L | [ |
| 苯甲醛 | BldR/MarR | 3F3X | 全细胞平台 | eGFP | ~µmol/L | [ |
| 3-羟基苯甲酸 | MobR/MarR | - | 无细胞平台 | 3WJdB荧光适配体/ | - | [ |
| 苯亚胂酸盐 | ArsR/ArsR | 6J05 | 全细胞平台 | GFP | 1 µmol/L | [ |
| 丙酮酸盐 | PdhR/GntR | - | 全细胞平台 | yEGFP | ~ fmol/L | [ |
| 抗生素 Antibiotic | 转录因子/家族 TFs/Families | PDB | 反应平台 Reaction platform | 报告元件 Report component | 检测限 Limit of detection | 参考文献 Reference |
|---|---|---|---|---|---|---|
| 四环素 | TetR/TetR | 1QPI | 限制性内切酶 | 荧光探针 | 25 nmol/L | [ |
| 全细胞平台 | EGFP和mCherry | 0.1 ng/mL | [ | |||
| aTF-NAST | RPA | 0.005 nmol/L | [ | |||
| 无细胞平台 | 荧光适配体 | ~125 nmol/L | [ | |||
| 无细胞平台 | G-四链体 | 18.08 ng/mL | [ | |||
| 无细胞平台 | 荧光适配体 | 10 nmol/L | [ | |||
| 无细胞平台 | CRISPR-Cas12a | - | [ | |||
| 无细胞平台 | 荧光素酶 | 45 nmol/L | [ | |||
| 脱水四环素 | QD-FRET | 光致发光 | 6 nmol/L | [ | ||
| QD-FRET | 光致发光 | 80 nmol/L | [ | |||
| 强力霉素 | 全细胞平台 | yEmRFP | 0.3 µg/mL | [ | ||
| 无细胞平台 | 荧光适配体 | ~0.2 µmol/L | [ | |||
| 放线菌素 | ActR/TetR | 3B6C | 全细胞平台 | - | [ | |
| 金霉素 | MphR/MphR | 3FRQ | 无细胞平台 | 荧光适配体 | ~125 nmol/L | [ |
| 放线菌素 | 全细胞平台 | GFP | - | [ | ||
| 红霉素 | 全细胞平台 | GFP | 13 nmol/L | [ | ||
| 无细胞平台 | 荧光适配体 | ~2.5 µmol/L | [ | |||
| 无细胞平台 | 荧光素酶 | 7.3 nmol/L | [ | |||
| 罗红霉素 | 无细胞平台 | 荧光适配体 | ~2.5 µmol/L | [ | ||
| 阿奇霉素 | 荧光适配体 | 荧光适配体 | ~2.5 µmol/L | [ | ||
| 叠氮红霉素 | 全细胞平台 | 荧光素酶 | - | [ | ||
| 竹桃霉素 | 全细胞平台 | 荧光素酶 | - | [ | ||
| 苦霉素 | 全细胞平台 | 荧光素酶 | - | [ | ||
| 克拉霉素 | MphR/MphR | 6U18 | 全细胞平台 | GFP | 1 µmol/L | [ |
| 无细胞平台 | 荧光适配体 | ~2.5 µmol/L | [ | |||
| 土霉素 | CtcS/MarR | 无细胞平台 | 荧光适配体 | ~125 nmol/L | [ | |
| 美罗培南 | AmpR/LysR | 全细胞平台 | mCherry | 8 pg/mL | [ | |
| 亚胺培南 | 全细胞平台 | mCherry | 40 pg/mL | [ |
Table 3 Detection of antibiotic by transcription factor-based biosensors
| 抗生素 Antibiotic | 转录因子/家族 TFs/Families | PDB | 反应平台 Reaction platform | 报告元件 Report component | 检测限 Limit of detection | 参考文献 Reference |
|---|---|---|---|---|---|---|
| 四环素 | TetR/TetR | 1QPI | 限制性内切酶 | 荧光探针 | 25 nmol/L | [ |
| 全细胞平台 | EGFP和mCherry | 0.1 ng/mL | [ | |||
| aTF-NAST | RPA | 0.005 nmol/L | [ | |||
| 无细胞平台 | 荧光适配体 | ~125 nmol/L | [ | |||
| 无细胞平台 | G-四链体 | 18.08 ng/mL | [ | |||
| 无细胞平台 | 荧光适配体 | 10 nmol/L | [ | |||
| 无细胞平台 | CRISPR-Cas12a | - | [ | |||
| 无细胞平台 | 荧光素酶 | 45 nmol/L | [ | |||
| 脱水四环素 | QD-FRET | 光致发光 | 6 nmol/L | [ | ||
| QD-FRET | 光致发光 | 80 nmol/L | [ | |||
| 强力霉素 | 全细胞平台 | yEmRFP | 0.3 µg/mL | [ | ||
| 无细胞平台 | 荧光适配体 | ~0.2 µmol/L | [ | |||
| 放线菌素 | ActR/TetR | 3B6C | 全细胞平台 | - | [ | |
| 金霉素 | MphR/MphR | 3FRQ | 无细胞平台 | 荧光适配体 | ~125 nmol/L | [ |
| 放线菌素 | 全细胞平台 | GFP | - | [ | ||
| 红霉素 | 全细胞平台 | GFP | 13 nmol/L | [ | ||
| 无细胞平台 | 荧光适配体 | ~2.5 µmol/L | [ | |||
| 无细胞平台 | 荧光素酶 | 7.3 nmol/L | [ | |||
| 罗红霉素 | 无细胞平台 | 荧光适配体 | ~2.5 µmol/L | [ | ||
| 阿奇霉素 | 荧光适配体 | 荧光适配体 | ~2.5 µmol/L | [ | ||
| 叠氮红霉素 | 全细胞平台 | 荧光素酶 | - | [ | ||
| 竹桃霉素 | 全细胞平台 | 荧光素酶 | - | [ | ||
| 苦霉素 | 全细胞平台 | 荧光素酶 | - | [ | ||
| 克拉霉素 | MphR/MphR | 6U18 | 全细胞平台 | GFP | 1 µmol/L | [ |
| 无细胞平台 | 荧光适配体 | ~2.5 µmol/L | [ | |||
| 土霉素 | CtcS/MarR | 无细胞平台 | 荧光适配体 | ~125 nmol/L | [ | |
| 美罗培南 | AmpR/LysR | 全细胞平台 | mCherry | 8 pg/mL | [ | |
| 亚胺培南 | 全细胞平台 | mCherry | 40 pg/mL | [ |
| [1] |
杨克恭. 细菌有转录因子吗? —“转录因子” 概念的形成和发展浅析[J]. 中国生物化学与分子生物学报, 2021, 37(6): 691-696.
doi: 10.13865/j.cnki.cjbmb.2021.05.1130 |
| Yang KG. Do bacteria have the transcription factors? The formation and development of the concept of “transcription factor”[J]. Chin J Biochem Mol Biol, 2021, 37(6): 691-696. | |
| [2] | 卜恺璇, 周翠霞, 路福平, 等. 细菌转录起始调控机制[J]. 中国生物工程杂志, 2021, 41(11): 89-99. |
| Bu KX, Zhou CX, Lu FP, et al. Research on the regulation mechanism of bacterial transcription initiation[J]. China Biotechnol, 2021, 41(11): 89-99. | |
| [3] |
杨璐, 吴楠, 白茸茸, 等. 基因回路型全细胞微生物传感器的设计、优化与应用[J]. 合成生物学, 2022, 3(6): 1061-1080.
doi: 10.12211/2096-8280.2021-021 |
| Yang L, Wu N, Bai RR, et al. Design, optimization and application of whole-cell microbial biosensors with engineered genetic circuits[J]. Synth Biol J, 2022, 3(6): 1061-1080. | |
| [4] | 赵晓蕊, 陈慧芳, 雷春阳, 等. 无细胞生物传感技术在环境污染物检测方面的应用进展[J]. 分析化学, 2023, 51(8): 1223-1231. |
| Zhao XR, Chen HF, Lei CY, et al. Advances in application of cell-free biosensing technologies for detection of environmental pollutant[J]. Chin J Anal Chem, 2023, 51(8): 1223-1231. | |
| [5] | Gu YQ, Fan F, Liu Y, et al. Cell-free protein synthesis system for bioanalysis: advances in methods and applications[J]. Trac Trends Anal Chem, 2023, 161: 117015. |
| [6] | Li SS, Li ZL, Tan GY, et al. In vitro allosteric transcription factor-based biosensing[J]. Trends Biotechnol, 2023, 41(8): 1080-1095. |
| [7] | Li SS, Zhou L, Yao YP, et al. A platform for the development of novel biosensors by configuring allosteric transcription factor recognition with amplified luminescent proximity homogeneous assays[J]. Chem Commun, 2017, 53(1): 99-102. |
| [8] | Nguyen TT, Chern M, Baer RC, et al. A Förster resonance energy transfer-based ratiometric sensor with the allosteric transcription factor TetR[J]. Small, 2020, 16(17): e1907522. |
| [9] | Yao YP, Li SS, Cao JQ, et al. Development of small molecule biosensors by coupling the recognition of the bacterial allosteric transcription factor with isothermal strand displacement amplification[J]. Chem Commun, 2018, 54(38): 4774-4777. |
| [10] |
Yao YP, Li SS, Cao JQ, et al. A novel signal transduction system for development of uric acid biosensors[J]. Appl Microbiol Biotechnol, 2018, 102(17): 7489-7497.
doi: 10.1007/s00253-018-9056-8 pmid: 29961098 |
| [11] | Cao JQ, Yao YP, Fan KQ, et al. Harnessing a previously unidentified capability of bacterial allosteric transcription factors for sensing diverse small molecules in vitro[J]. Sci Adv, 2018, 4(11): eaau4602. |
| [12] | Iwasaki RS, Batey RT. SPRINT: a Cas13a-based platform for detection of small molecules[J]. Nucleic Acids Res, 2020, 48(17): e101. |
| [13] |
Eddy SR. Computational genomics of noncoding RNA genes[J]. Cell, 2002, 109(2): 137-140.
pmid: 12007398 |
| [14] | Gajiwala KS, Burley SK. Winged helix proteins[J]. Curr Opin Struct Biol, 2000, 10(1): 110-116. |
| [15] |
Schreiter ER, Drennan CL. Ribbon-helix-helix transcription factors: variations on a theme[J]. Nat Rev Microbiol, 2007, 5(9): 710-720.
doi: 10.1038/nrmicro1717 pmid: 17676053 |
| [16] |
Yu QK, Ren KW, You MX. Genetically encoded RNA nanodevices for cellular imaging and regulation[J]. Nanoscale, 2021, 13(17): 7988-8003.
doi: 10.1039/d0nr08301a pmid: 33885099 |
| [17] |
Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein[J]. Science, 2011, 333(6042): 642-646.
doi: 10.1126/science.1207339 pmid: 21798953 |
| [18] | Jung JK, Alam KK, Verosloff MS, et al. Cell-free biosensors for rapid detection of water contaminants[J]. Nat Biotechnol, 2020, 38(12): 1451-1459. |
| [19] | Qiu CX, Zhai HT, Hou J. Biosensors design in yeast and applications in metabolic engineering[J]. FEMS Yeast Res, 2019, 19(8): foz082. |
| [20] |
Mukherjee K, Bhattacharyya S, Peralta-Yahya P. GPCR-based chemical biosensors for medium-chain fatty acids[J]. ACS Synth Biol, 2015, 4(12): 1261-1269.
doi: 10.1021/sb500365m pmid: 25992593 |
| [21] |
Dong CY, Ly C, Dunlap LE, et al. Psychedelic-inspired drug discovery using an engineered biosensor[J]. Cell, 2021, 184(10): 2779-2792.e18.
doi: 10.1016/j.cell.2021.03.043 pmid: 33915107 |
| [22] | Dhakal S, Macreadie I. The use of yeast in biosensing[J]. Microorganisms, 2022, 10(9): 1772. |
| [23] | Wahid E, Ocheja OB, Marsili E, et al. Biological and technical challenges for implementation of yeast-based biosensors[J]. Microb Biotechnol, 2023, 16(1): 54-66. |
| [24] | Cerminati S, Soncini FC, Checa SK. A sensitive whole-cell biosensor for the simultaneous detection of a broad-spectrum of toxic heavy metal ions[J]. Chem Commun, 2015, 51(27): 5917-5920. |
| [25] | Mendoza JI, Soncini FC, Checa SK. Engineering of a Au-sensor to develop a Hg-specific, sensitive and robust whole-cell biosensor for on-site water monitoring[J]. Chem Commun, 2020, 56(48): 6590-6593. |
| [26] |
Chong HQ, Ching CB. Development of colorimetric-based whole-cell biosensor for organophosphorus compounds by engineering transcription regulator DmpR[J]. ACS Synth Biol, 2016, 5(11): 1290-1298.
pmid: 27346389 |
| [27] | Jha RK, Bingen JM, Johnson CW, et al. A protocatechuate biosensor for Pseudomonas putida KT2440 via promoter and protein evolution[J]. Metab Eng Commun, 2018, 6: 33-38. |
| [28] | Pu W, Chen JZ, Liu P, et al. Directed evolution of linker helix as an efficient strategy for engineering LysR-type transcriptional regulators as whole-cell biosensors[J]. Biosens Bioelectron, 2023, 222: 115004. |
| [29] | Zhang P, Yang MJ, Lan JJ, et al. Water quality degradation due to heavy metal contamination: health impacts and eco-friendly approaches for heavy metal remediation[J]. Toxics, 2023, 11(10): 828. |
| [30] | 朱振宇, 华垚堃, 胡婷婷, 等. 微生物金属响应蛋白研究进展[J]. 微生物学通报, 2018, 45(8): 1794-1803. |
| Zhu ZY, Hua YK, Hu TT, et al. Advances in microbial metal response proteins[J]. Microbiol China, 2018, 45(8): 1794-1803. | |
| [31] | Guo Y, Hui CY, Liu LS, et al. Development of a bioavailable Hg(II)sensing system based on MerR-regulated visual pigment biosynthesis[J]. Sci Rep, 2021, 11(1): 13516. |
| [32] | Guo MZ, Wang JL, Du RX, et al. A test strip platform based on a whole-cell microbial biosensor for simultaneous on-site detection of total inorganic mercury pollutants in cosmetics without the need for predigestion[J]. Biosens Bioelectron, 2020, 150: 111899. |
| [33] | Wang D, Zheng YN, Xu LN, et al. Engineered cells for selective detection and remediation of Hg2+ based on transcription factor MerR regulated cell surface displayed systems[J]. Biochem Eng J, 2019, 150: 107289. |
| [34] |
Bereza-Malcolm L, Aracic S, Kannan RB, et al. Functional characterization of Gram-negative bacteria from different Genera as multiplex cadmium biosensors[J]. Biosens Bioelectron, 2017, 94: 380-387.
doi: S0956-5663(17)30180-X pmid: 28319906 |
| [35] | Tang TC, Tham E, Liu XY, et al. Hydrogel-based biocontainment of bacteria for continuous sensing and computation[J]. Nat Chem Biol, 2021, 17(6): 724-731. |
| [36] |
Wang BJ, Barahona M, Buck M. A modular cell-based biosensor using engineered genetic logic circuits to detect and integrate multiple environmental signals[J]. Biosens Bioelectron, 2013, 40(1): 368-376.
doi: 10.1016/j.bios.2012.08.011 pmid: 22981411 |
| [37] | Cayron J, Prudent E, Escoffier C, et al. Pushing the limits of nickel detection to nanomolar range using a set of engineered bioluminescent Escherichia coli[J]. Environ Sci Pollut Res Int, 2017, 24(1): 4-14. |
| [38] |
Kim HJ, Jeong H, Lee SJ. Synthetic biology for microbial heavy metal biosensors[J]. Anal Bioanal Chem, 2018, 410(4): 1191-1203.
doi: 10.1007/s00216-017-0751-6 pmid: 29184994 |
| [39] | Zhao XY, Dong T. A microfluidic device for continuous sensing of systemic acute toxicants in drinking water[J]. Int J Environ Res Public Health, 2013, 10(12): 6748-6763. |
| [40] | Zhang YK, Zhao C, Bi HX, et al. A cell-free paper-based biosensor dependent on allosteric transcription factors(aTFs)for on-site detection of harmful metals Hg2+ and Pb2+ in water[J]. J Hazard Mater, 2022, 438: 129499. |
| [41] | Lin PH, Tsai ST, Chang YC, et al. Harnessing split fluorescent proteins in modular protein logic for advanced whole-cell detection[J]. Anal Chim Acta, 2023, 1275: 341593. |
| [42] | Elcin E, Öktem HA. Inorganic cadmium detection using a fluorescent whole-cell bacterial bioreporter[J]. Anal Lett, 2020, 53(17): 2715-2733. |
| [43] | Kim H, Lee W, Yoon Y. Heavy metal(loid)biosensor based on split-enhanced green fluorescent protein: development and characterization[J]. Appl Microbiol Biotechnol, 2019, 103(15): 6345-6352. |
| [44] | Hou QH, Ma AZ, Wang T, et al. Detection of bioavailable cadmium, lead, and arsenic in polluted soil by tailored multiple Escherichia coli whole-cell sensor set[J]. Anal Bioanal Chem, 2015, 407(22): 6865-6871. |
| [45] |
Kim HJ, Lim JW, Jeong H, et al. Development of a highly specific and sensitive cadmium and lead microbial biosensor using synthetic CadC-T7 genetic circuitry[J]. Biosens Bioelectron, 2016, 79: 701-708.
doi: 10.1016/j.bios.2015.12.101 pmid: 26773374 |
| [46] | Kumar S, Verma N, Singh AK. Development of cadmium specific recombinant biosensor and its application in milk samples[J]. Sens Actuat B Chem, 2017, 240: 248-254. |
| [47] | Jia XQ, Liu T, Ma YB, et al. Construction of cadmium whole-cell biosensors and circuit amplification[J]. Appl Microbiol Biotechnol, 2021, 105(13): 5689-5699. |
| [48] | Fernández M, Morel B, Ramos JL, et al. Paralogous regulators ArsR1 and ArsR2 of Pseudomonas putida KT2440 as a basis for arsenic biosensor development[J]. Appl Environ Microbiol, 2016, 82(14): 4133-4144. |
| [49] | Chen PH, Lin C, Guo KH, et al. Development of a pigment-based whole-cell biosensor for the analysis of environmental copper[J]. RSC Adv, 2017, 7(47): 29302-29305. |
| [50] | Kang Y, Lee W, Kim S, et al. Enhancing the copper-sensing capability of Escherichia coli-based whole-cell bioreporters by genetic engineering[J]. Appl Microbiol Biotechnol, 2018, 102(3): 1513-1521. |
| [51] |
Date A, Pasini P, Daunert S. Construction of spores for portable bacterial whole-cell biosensing systems[J]. Anal Chem, 2007, 79(24): 9391-9397.
pmid: 18020369 |
| [52] | Yoon Y, Kim S, Chae Y, et al. Use of tunable whole-cell bioreporters to assess bioavailable cadmium and remediation performance in soils[J]. PLoS One, 2016, 11(5): e0154506. |
| [53] |
Yoon Y, Kim S, Chae Y, et al. Simultaneous detection of bioavailable arsenic and cadmium in contaminated soils using dual-sensing bioreporters[J]. Appl Microbiol Biotechnol, 2016, 100(8): 3713-3722.
doi: 10.1007/s00253-016-7338-6 pmid: 26852408 |
| [54] |
Ghataora JS, Gebhard S, Reeksting BJ. Chimeric MerR-family regulators and logic elements for the design of metal sensitive genetic circuits in Bacillus subtilis[J]. ACS Synth Biol, 2023, 12(3): 735-749.
doi: 10.1021/acssynbio.2c00545 pmid: 36629785 |
| [55] | Hui CY, Guo Y, Li LM, et al. Indigoidine biosynthesis triggered by the heavy metal-responsive transcription regulator: a visual whole-cell biosensor[J]. Appl Microbiol Biotechnol, 2021, 105(14-15): 6087-6102. |
| [56] | Hui CY, Guo Y, Zhu DL, et al. Metabolic engineering of the violacein biosynthetic pathway toward a low-cost, minimal-equipment lead biosensor[J]. Biosens Bioelectron, 2022, 214: 114531. |
| [57] |
Lee W, Kim H, Jang G, et al. Antimony sensing whole-cell bioreporters derived from ArsR genetic engineering[J]. Appl Microbiol Biotechnol, 2020, 104(6): 2691-2699.
doi: 10.1007/s00253-020-10413-5 pmid: 32002600 |
| [58] |
Muir T, Michalek JE, Palmer RF. Determination of safe levels of persistent organic pollutants in toxicology and epidemiology[J]. Rev Environ Health, 2022, 38(3): 401-408.
doi: 10.1515/reveh-2021-0105 pmid: 35506713 |
| [59] | 郭庆伟, 王倩, 张海东, 等. 新污染物检测技术研究进展[J]. 化学通报, 2024, 87(1): 78-85. |
| Guo QW, Wang Q, Zhang HD, et al. Research progress in detection technology of emerging pollutants[J]. Chemistry, 2024, 87(1): 78-85. | |
| [60] | Sun SW, Peng KL, Sun S, et al. Engineering modular and highly sensitive cell-based biosensors for aromatic contaminant monitoring and high-throughput enzyme screening[J]. ACS Synth Biol, 2023, 12(3): 877-891. |
| [61] | Darjee SM, Modi KM, Panchal U, et al. Highly selective and sensitive fluorescent sensor: Thiacalix[4]arene-1-naphthalene carboxylate for Zn2+ ions[J]. J Mol Struct, 2017, 1133: 1-8. |
| [62] |
Sun YJ, Zhao XH, Zhang DY, et al. New naphthalene whole-cell bioreporter for measuring and assessing naphthalene in polycyclic aromatic hydrocarbons contaminated site[J]. Chemosphere, 2017, 186: 510-518.
doi: S0045-6535(17)31248-1 pmid: 28810221 |
| [63] | Roy R, Ray S, Chowdhury A, et al. Tunable multiplexed whole-cell biosensors as environmental diagnostics for ppb-level detection of aromatic pollutants[J]. ACS Sens, 2021, 6(5): 1933-1939. |
| [64] | Juárez JF, Lecube-Azpeitia B, Brown SL, et al. Biosensor libraries harness large classes of binding domains for construction of allosteric transcriptional regulators[J]. Nat Commun, 2018, 9(1): 3101. |
| [65] | Voyvodic PL, Pandi A, Koch M, et al. Plug-and-play metabolic transducers expand the chemical detection space of cell-free biosensors[J]. Nat Commun, 2019, 10(1): 1697. |
| [66] |
Zhang P, Feng HB, Yang JZ, et al. Detection of inorganic ions and organic molecules with cell-free biosensing systems[J]. J Biotechnol, 2019, 300: 78-86.
doi: S0168-1656(19)30175-0 pmid: 31141711 |
| [67] | Ray S, Panjikar S, Anand R. Design of protein-based biosensors for selective detection of benzene groups of pollutants[J]. ACS Sens, 2018, 3(9): 1632-1638. |
| [68] | Campos VL, Zaror CA, Mondaca MA. Detection of chlorinated phenols in kraft pulp bleaching effluents using DmpR mutant strains[J]. Bull Environ Contam Toxicol, 2004, 73(4): 666-673. |
| [69] |
Behzadian F, Barjeste H, Hosseinkhani S, et al. Construction and characterization of Escherichia coli whole-cell biosensors for toluene and related compounds[J]. Curr Microbiol, 2011, 62(2): 690-696.
doi: 10.1007/s00284-010-9764-5 pmid: 20872219 |
| [70] |
Garmendia J, de las Heras A, Galvão TC, et al. Tracing explosives in soil with transcriptional regulators of Pseudomonas putida evolved for responding to nitrotoluenes[J]. Microb Biotechnol, 2008, 1(3): 236-246.
doi: 10.1111/j.1751-7915.2008.00027.x pmid: 21261843 |
| [71] | de Las Heras A, de Lorenzo V. Cooperative amino acid changes shift the response of the σ54-dependent regulator XylR from natural m-xylene towards xenobiotic 2, 4-dinitrotoluene[J]. Mol Microbiol, 2011, 79(5): 1248-1259. |
| [72] |
Turner K, Xu SF, Pasini P, et al. Hydroxylated polychlorinated biphenyl detection based on a genetically engineered bioluminescent whole-cell sensing system[J]. Anal Chem, 2007, 79(15): 5740-5745.
pmid: 17602671 |
| [73] | Werlen C, Jaspers MCM, van der Meer JR. van der Meer JR. Measurement of biologically available naphthalene in gas and aqueous phases by use of a Pseudomonas putida biosensor[J]. Appl Environ Microbiol, 2004, 70(1): 43-51. |
| [74] |
Nasr MA, Timmins LR, Martin VJJ, et al. A versatile transcription factor biosensor system responsive to multiple aromatic and indole inducers[J]. ACS Synth Biol, 2022, 11(4): 1692-1698.
doi: 10.1021/acssynbio.2c00063 pmid: 35316041 |
| [75] | Asemoloye MD, Marchisio MA. Synthetic metabolic transducers in Saccharomyces cerevisiae as sensors for aromatic permeant acids and bioreporters of hydrocarbon metabolism[J]. Biosens Bioelectron, 2023, 220: 114897. |
| [76] |
Fiorentino G, Ronca R, Bartolucci S. A novel E. coli biosensor for detecting aromatic aldehydes based on a responsive inducible archaeal promoter fused to the green fluorescent protein[J]. Appl Microbiol Biotechnol, 2009, 82(1): 67-77.
doi: 10.1007/s00253-008-1771-0 pmid: 18998120 |
| [77] | Chen J, Sun S, Li CZ, et al. Biosensor for organoarsenical herbicides and growth promoters[J]. Environ Sci Technol, 2014, 48(2): 1141-1147. |
| [78] | Anomaly J. Harm to others: the social cost of antibiotics in agriculture[J]. J Agric Environ Ethics, 2009, 22(5): 423-435. |
| [79] |
Founou LL, Founou RC, Essack SY. Antibiotic resistance in the food chain: a developing country-perspective[J]. Front Microbiol, 2016, 7: 1881.
pmid: 27933044 |
| [80] | Chern M, Garden PM, Baer RC, et al. Transcription factor based small-molecule sensing with a rapid cell phone enabled fluorescent bead assay[J]. Angew Chem Int Ed Engl, 2020, 59(48): 21597-21602. |
| [81] | Zhang R, Wang Y, Deng HF, et al. Fast and bioluminescent detection of antibiotic contaminants by on-demand transcription of RNA scaffold arrays[J]. Anal Chim Acta, 2023, 1273: 341538. |
| [82] | Richter MF, Drown BS, Riley AP, et al. Predictive compound accumulation rules yield a broad-spectrum antibiotic[J]. Nature, 2017, 545(7654): 299-304. |
| [83] |
Miller CA, Ho JM, Parks SE, et al. Macrolide biosensor optimization through cellular substrate sequestration[J]. ACS Synth Biol, 2021, 10(2): 258-264.
doi: 10.1021/acssynbio.0c00572 pmid: 33555859 |
| [84] | Li SL, Chen DD, Liu ZQ, et al. Directed evolution of TetR for constructing sensitive and broad-spectrum tetracycline antibiotics whole-cell biosensor[J]. J Hazard Mater, 2023, 460: 132311. |
| [85] |
Rodríguez-Serrano AF, Hsing IM. Allosteric regulation of DNA circuits enables minimal and rapid biosensors of small molecules[J]. ACS Synth Biol, 2021, 10(2): 371-378.
doi: 10.1021/acssynbio.0c00545 pmid: 33481567 |
| [86] | Ullrich T, Weirich S, Jeltsch A. Development of an epigenetic tetracycline sensor system based on DNA methylation[J]. PLoS One, 2020, 15(5): e0232701. |
| [87] | Liu RN, Liu X, Yang H, et al. A cell-free biosensor based on strand displacement amplification and hybridization chain reaction for fluorescence detection of tetracycline[J]. Microchem J, 2023, 185: 108239. |
| [88] | Bi HX, Zhao C, Zhang YK, et al. IVT cell-free biosensors for tetracycline and macrolide detection based on allosteric transcription factors(aTFs)[J]. Anal Methods, 2022, 14(44): 4545-4554. |
| [89] |
Mahas A, Wang QC, Marsic T, et al. Development of Cas12a-based cell-free small-molecule biosensors via allosteric regulation of CRISPR array expression[J]. Anal Chem, 2022, 94(11): 4617-4626.
doi: 10.1021/acs.analchem.1c04332 pmid: 35266687 |
| [90] | Chen MF, Nguyen TT, Varongchayakul N, et al. Surface immobilized nucleic acid-transcription factor quantum dots for biosensing[J]. Adv Healthc Mater, 2020, 9(17): e2000403. |
| [91] | Miller RA, Brown G, Barron E, et al. Development of a paper-immobilized yeast biosensor for the detection of physiological concentrations of doxycycline in technology-limited settings[J]. Anal Methods, 2020, 12(16): 2123-2132. |
| [92] | Ahn SK, Tahlan K, Yu Z, et al. Investigation of transcription repression and small-molecule responsiveness by TetR-like transcription factors using a heterologous Escherichia coli-based assay[J]. J Bacteriol, 2007, 189(18): 6655-6664. |
| [93] |
Kasey CM, Zerrad M, Li YW, et al. Development of transcription factor-based designer macrolide biosensors for metabolic engineering and synthetic biology[J]. ACS Synth Biol, 2018, 7(1): 227-239.
doi: 10.1021/acssynbio.7b00287 pmid: 28950701 |
| [94] | Möhrle V, Stadler M, Eberz G. Biosensor-guided screening for macrolides[J]. Anal Bioanal Chem, 2007, 388(5/6): 1117-1125. |
| [95] |
Li YW, Reed M, Wright HT, et al. Development of genetically encoded biosensors for reporting the methyltransferase-dependent biosynthesis of semisynthetic macrolide antibiotics[J]. ACS Synth Biol, 2021, 10(10): 2520-2531.
doi: 10.1021/acssynbio.1c00151 pmid: 34546703 |
| [96] | Higuera-Llantén S, Alcalde-Rico M, Vasquez-Ponce F, et al. A whole-cell hypersensitive biosensor for beta-lactams based on the AmpR-AmpC regulatory circuit from the Antarctic Pseudomonas sp. IB20[J]. Microb Biotechnol, 2024, 17(1): e14385. |
| [97] | Liu CJ, Yu H, Zhang BC, et al. Engineering whole-cell microbial biosensors: design principles and applications in monitoring and treatment of heavy metals and organic pollutants[J]. Biotechnol Adv, 2022, 60: 108019. |
| [1] | HU Ya-dan, WU Guo-qiang, LIU Chen, WEI Ming. Roles of MYB Transcription Factor in Regulating the Responses of Plants to Stress [J]. Biotechnology Bulletin, 2024, 40(6): 5-22. |
| [2] | WANG Di ZHANG Xiao-yu SONG Yu-xin ZHENG Dong-ran TIAN Jing LI Yu-hua WANG Yu WU Hao. Advances in the Molecular Mechanisms of Plant Tissue Culture and Regeneration Regulated by Totipotency-related Transcription Factors [J]. Biotechnology Bulletin, 2024, 40(6): 23-33. |
| [3] | GUO Chun, SONG Gui-mei, YAN Yan, DI Peng, WANG Ying-ping. Genome Wide Identification and Expression Analysis of the bZIP Gene Family in Panax quinquefolius [J]. Biotechnology Bulletin, 2024, 40(4): 167-178. |
| [4] | CHEN Zhi-min, LI Cui, WEI Ji-tian, LI Xin-ran, LIU Yi, GUO Qiang. Research Progress in the Regulation of Chlorogenic Acid Biosynthesis and Its Application [J]. Biotechnology Bulletin, 2024, 40(1): 57-71. |
| [5] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [6] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [7] | CHEN Yong, LI Ya-xin, WANG Ya-xuan, LIANG Lu-jie, FENG Si-yuan, Tian Guo-bao. Research Progress in the Molecular Mechanism of MCR-1 Mediated Polymyxin Resistance [J]. Biotechnology Bulletin, 2023, 39(6): 102-108. |
| [8] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [9] | ZHANG Xin-bo, CUI Hao-liang, SHI Pei-hua, GAO Jin-chun, ZHAO Shun-ran, TAO Chen-yu. Research Progress in Low-input Chromatin Immunoprecipitation Assay [J]. Biotechnology Bulletin, 2023, 39(4): 227-235. |
| [10] | ZHAO Meng-liang, GUO Yi-ting, REN Yan-jing. Identification and Analysis of WRKY Transcription Factor Family Genes in Helianthus tuberosus [J]. Biotechnology Bulletin, 2023, 39(2): 116-125. |
| [11] | HAN Fang-ying, HU Xin, WANG Nan-nan, XIE Yu-hong, WANG Xiao-yan, ZHU Qiang. Research Progress in Response of DREBs to Abiotic Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(11): 86-98. |
| [12] | FENG Ce-ting, JIANG Lyu, LIU Xing-ying, LUO Le, PAN Hui-tang, ZHANG Qi-xiang, YU Chao. Identification of the NAC Gene Family in Rosa persica and Response Analysis Under Drought Stress [J]. Biotechnology Bulletin, 2023, 39(11): 283-296. |
| [13] | CHEN Hao-ting, ZHANG Yu-jing, LIU Jie, DAI Ze-min, LIU Wei, SHI Yu, ZHANG Yi, LI Tian-lai. Functional Analysis of WRKY6 Gene in Tomato Under Low-phosphorus Stress [J]. Biotechnology Bulletin, 2023, 39(10): 136-147. |
| [14] | LI Ren-han, ZHANG Le-le, LIU Chun-li, LIU Xiu-xia, BAI Zhong-hu, YANG Yan-kun, LI Ye. Development of an L-tryptophan Biosensor Based on the Violacein Biosynthesis Pathway [J]. Biotechnology Bulletin, 2023, 39(10): 80-92. |
| [15] | CHEN Xiao-lin, LIU Yang-er, XU Wen-tao, GUO Ming-zhang, LIU Hui-lin. Application of Synthetic Biology Based Whole-cell Biosensor Technology in the Rapid Detection of Food Safety [J]. Biotechnology Bulletin, 2023, 39(1): 137-149. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||