








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (7): 28-42.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1065
Previous Articles Next Articles
WANG Qing( ), NI Er-dong, WANG Qiu-shuang, QIN Dan-dan, FANG Kai-xing, LI Hong-jian, JIANG Xiao-hui, LI Bo, PAN Chen-dong, WU Hua-ling(
), NI Er-dong, WANG Qiu-shuang, QIN Dan-dan, FANG Kai-xing, LI Hong-jian, JIANG Xiao-hui, LI Bo, PAN Chen-dong, WU Hua-ling( )
)
Received:2023-11-10
Online:2024-07-26
Published:2024-07-30
Contact:
WU Hua-ling
E-mail:rosewangqing@126.com;wuhualing@163.com
WANG Qing, NI Er-dong, WANG Qiu-shuang, QIN Dan-dan, FANG Kai-xing, LI Hong-jian, JIANG Xiao-hui, LI Bo, PAN Chen-dong, WU Hua-ling. Research Progress of UDP-glycosyltransferase Related to Anthocyanin Modification in Plants[J]. Biotechnology Bulletin, 2024, 40(7): 28-42.

Fig. 1 PSPG-box consensus sequence of UDP-glycosyltransferases in plants Highly conserved amino acids are shaded in blue(identity>50%)or red(identity>80%)
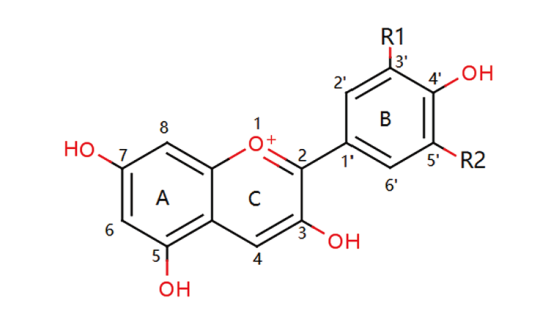
Fig. 3 Chemical structures of anthocyanidins Pelargonidin: R1=H, R2=H. Cyanidin:R1=OH, R2=H. Peonidin: R1=OCH3, R2=H. Delphindin: R1=OH, R2=OH. Petunidin: R1=OCH3, R2=OH. Malvidin: R1=OCH3, R2=OCH3
| 物种 Species | 基因 Gene | GenBank登录号 GenBank accession No. | 糖供体 Sugar donor | 底物 Substrate | 产物 Product | 参考文献 Reference |
|---|---|---|---|---|---|---|
| 矮牵牛 | PGT8 | - | UDP-葡萄糖 | 天竺葵素,矢车菊色素,飞燕草素 | 天竺葵素 3-O-葡萄糖苷,矢车菊色素 3-O-葡萄糖苷,飞燕草素 3-O-葡萄糖苷 | [ |
| 矮牵牛 | PH1 | - | UDP-葡萄糖 | 飞燕草素 3-(p-酰基)-芸香糖苷 | 飞燕草素 3-(p-酰基)-芸香糖-5-O-葡萄糖苷 | [ |
| 拟南芥 | UGT78D2 | AT5G17050 | UDP-葡萄糖 | 矢车菊色素,天竺葵素,飞燕草色素 | 矢车菊色素 3-O-葡萄糖苷,天竺葵素 3-O-葡萄糖苷,飞燕草色素 3-O-葡萄糖苷 | [ |
| 拟南芥 | UGT79B2 | AT4G27560 | UDP-鼠李糖 | 矢车菊色素,矢车菊色素 3-O-葡萄糖苷 | 矢车菊色素 3-O-鼠李糖苷,矢车菊色素 3-O-葡萄糖-鼠李糖苷 | [ |
| 拟南芥 | UGT79B3 | AT4G27570 | UDP-鼠李糖 | 矢车菊色素,矢车菊色素 3-O-葡萄糖苷 | 矢车菊色素 3-O-鼠李糖苷,矢车菊色素 3-O-葡萄糖-鼠李糖苷 | [ |
| 拟南芥 | UGT79B1 | AT5G54060 | UDP-木糖 | 矢车菊色素 3-O-葡萄糖苷 | 矢车菊色素 3-O-木糖(1-2)-葡萄糖苷 | [ |
| 猕猴桃 | F3GT1 | GU079683 | UDP-半乳糖 | 矢车菊色素 | 矢车菊色素 3-O-半乳糖苷 | [ |
| 猕猴桃 | F3GGT1 | FG404013 | UDP-木糖 | 矢车菊色素 3-O-半乳糖苷 | 矢车菊色素 3-O-木糖(1-2)-半乳糖苷 | [ |
| 猕猴桃 | AcUFGT3a | AYJ72756 | UDP-半乳糖 | 矢车菊色素 | 矢车菊色素 3-O-半乳糖苷 | [ |
| 猕猴桃 | AcUFGT6b | MH817045 | UDP-木糖 | 矢车菊色素 3-O-半乳糖苷 | 矢车菊色素 3-O-木糖-半乳糖苷 | [ |
| 桃 | PpUGT78A1 | - | UDP-半乳糖 | 矢车菊色素 | 矢车菊色素-3-半乳糖苷 | [ |
| 桃 | PpUGT78B | - | UDP-葡萄糖 | 矢车菊色素 | 矢车菊色素-3-葡萄糖苷 | [ |
| 桃 | PpUGT78A2 | - | UDP-葡萄糖 | 矢车菊色素 | 矢车菊色素-3-葡萄糖苷 | [ |
| 荔枝 | LcUFGT1 | KR704274 | UDP-葡萄糖 | 矢车菊色素 | 矢车菊色素 3-O-葡萄糖苷 | [ |
| 柿 | DkFGT | BAI40148 | UDP-半乳糖 | 矢车菊色素 | 矢车菊色素 3-O-半乳糖苷 | [ |
| 胎里红枣 | ZjUGT79B1 | - | UDP-鼠李糖 | 矢车菊色素 3-O-葡萄糖苷 | 矢车菊色素 3-O-芸香糖苷 | [ |
| 草莓 | FaGT1 | AAU09442 | UDP-葡萄糖 | 天竺葵色素,矢车菊色素,芍药色素 | 天竺葵色素 3-O-葡萄糖苷,矢车菊色素 3-O-葡萄糖苷,芍药色素 3-O-葡萄糖苷 | [ |
| 芒果 | rMiUFGalT3 | LC474861 | UDP-半乳糖 | 矢车菊色素,飞燕草色素,天竺葵色素,芍药色素,矮牵牛色素 | 矢车菊色素 3-O-半乳糖苷,飞燕草色素 3-O-半乳糖苷,天竺葵色素 3-O-半乳糖苷,芍药色素 3-O-半乳糖苷,矮牵牛色素 3-O-半乳糖苷 | [ |
| 葡萄 | VvGT1 | - | UDP-葡萄糖 | 矢车菊色素 | 矢车菊色素 3-O-葡萄糖苷 | [ |
| 红卷心菜 | UGT78D5 | LC536577.1 | UDP-葡萄糖 | 矢车菊色素 | 矢车菊色素 5-O-葡萄糖苷 | [ |
| 红卷心菜 | UGT79B45 | LC536579.1 | UDP-葡萄糖 | 矢车菊色素 3-O-葡萄糖苷 | 矢车菊色素 3-O-槐糖苷 | [ |
| 红卷心菜 | UGT79B44 | LC536578.1 | UDP-葡萄糖 | 矢车菊色素 3-O-葡萄糖苷 | 矢车菊色素 3-O-槐糖苷 | [ |
| 小苍兰 | Fh3GT1 | ADK75021.1 | UDP-葡萄糖 | 芍药色素,矮牵牛色素,矢车菊色素,飞燕草色素,天竺葵色素,锦葵色素 | 芍药色素 3-O-葡萄糖苷,矮牵牛色素 3-O-葡萄糖苷,矢车菊色素 3-O-葡萄糖苷,飞燕草色素 3-O-葡萄糖苷,天竺葵色素 3-O-葡萄糖苷,锦葵色素 3-O-葡萄糖苷 | [ |
| 小苍兰 | Fh3GT3 | - | UDP-葡萄糖 | 芍药色素,矮牵牛色素,矢车菊色素 | 芍药色素 3-O-葡萄糖苷,矮牵牛色素 3-O-葡萄糖苷,矢车菊色素 3-O-葡萄糖苷 | [ |
| 牵牛花 | 3GGT | AB192315 | UDP-鼠李糖 | 天竺葵色素 3-O-葡萄糖苷,矢车菊色素 3-O-葡萄糖苷,飞燕草色素 3-O-葡萄糖苷 | 天竺葵色素 3-O-芸香糖苷,矢车菊色素 3-O-芸香糖苷,飞燕草色素 3-O-芸香糖苷 | [ |
| 半边莲 | ABRT2 | LC131336 | UDP-鼠李糖 | 飞燕草色素 3-O-葡糖苷,矢车菊色素 3-O-葡萄糖苷,天竺葵色素 3-O-葡萄糖苷, | 飞燕草色素 3-O-芸香糖苷,矢车菊色素 3-O-芸香糖苷,天竺葵色素 3-O-芸香糖苷 | [ |
| 半边莲 | ABRT4 | LC131337 | UDP-鼠李糖 | 飞燕草色素 3-O-葡萄糖苷,矢车菊色素 3-O-葡萄糖苷,天竺葵色素 3 O-葡萄糖苷,锦葵色素 3-O-葡萄糖苷 | 飞燕草色素 3-O-芸香糖苷,矢车菊色素 3-O-芸香糖苷,天竺葵色素 3-O-芸香糖苷,锦葵色素 3-O-芸香糖苷 | [ |
| 紫芹菜 | AgUCGalT1 | - | UDP-半乳糖 | 矢车菊色素 | 矢车菊色素 3-O-半乳糖苷 | [ |
| 紫萝卜 | DcUCGalT1 | KP319022 | UDP-半乳糖 | 矢车菊色素 | 矢车菊色素 3-O-半乳糖苷 | [ |
| 玫瑰 | RhGT1 | AB201048 | UDP-葡萄糖 | 矢车菊色素,矢车菊色素 5-O-葡萄糖苷 | 矢车菊色素 5-O-葡萄糖苷,矢车菊色素 3,5-O-葡萄糖苷 | [ |
| 白杨 | PtUGT78L1 | - | UDP-半乳糖 | 矢车菊色素 | 矢车菊色素 3-O-半乳糖苷 | [ |
| 黑豆 | UGT78K1 | Glyma07g30180 | UDP-葡萄糖 | 矢车菊色素 | 矢车菊色素 3-O-葡萄糖苷 | [ |
| 龙胆草 | Gt5GT7 | AB363839 | UDP-葡萄糖 | 飞燕草素 3-O-葡萄糖苷,矢车菊色素 3-O-葡萄糖苷,天竺葵色素 3-O-葡萄糖苷,锦葵色素 3-O-葡萄糖苷 | 飞燕草素 3,5-O-葡萄糖苷,矢车菊色素 3,5-O-葡萄糖苷,天竺葵色素 3,5-O-葡萄糖苷,锦葵色素 3,5-O-葡萄糖苷 | [ |
| 紫薯 | Ib3GGT | KF056328 | UDP-葡萄糖 | 矢车菊色素 3-O-葡萄糖苷, 芍药色素 3-O-葡萄糖苷 | 矢车菊色素 3-O-槐糖苷,芍药色素 3-O-槐糖苷 | [ |
| 荷兰鸢尾花 | Ih3GT | AB161175 | UDP-葡萄糖 | 飞燕草素,锦葵色素,芍药色素,矢车菊色素,天竺葵色素 | 飞燕草素 3-O-葡萄糖苷,锦葵色素 3-O-葡萄糖苷,芍药色素 3-O-葡萄糖苷,矢车菊色素 3-O-葡萄糖苷,天竺葵色素 3-O-葡萄糖苷 | [ |
| 茶树 | CsUGT78A15 | - | UDP -半乳糖 | 矢车菊色素,飞燕草色素 | 矢车菊色素 3-O-半乳糖苷,飞燕草色素 3-O-半乳糖苷 | [ |
Table 1 Plant glycosyltransferases involved in anthocyanin modification by enzyme activity identification in vitro
| 物种 Species | 基因 Gene | GenBank登录号 GenBank accession No. | 糖供体 Sugar donor | 底物 Substrate | 产物 Product | 参考文献 Reference |
|---|---|---|---|---|---|---|
| 矮牵牛 | PGT8 | - | UDP-葡萄糖 | 天竺葵素,矢车菊色素,飞燕草素 | 天竺葵素 3-O-葡萄糖苷,矢车菊色素 3-O-葡萄糖苷,飞燕草素 3-O-葡萄糖苷 | [ |
| 矮牵牛 | PH1 | - | UDP-葡萄糖 | 飞燕草素 3-(p-酰基)-芸香糖苷 | 飞燕草素 3-(p-酰基)-芸香糖-5-O-葡萄糖苷 | [ |
| 拟南芥 | UGT78D2 | AT5G17050 | UDP-葡萄糖 | 矢车菊色素,天竺葵素,飞燕草色素 | 矢车菊色素 3-O-葡萄糖苷,天竺葵素 3-O-葡萄糖苷,飞燕草色素 3-O-葡萄糖苷 | [ |
| 拟南芥 | UGT79B2 | AT4G27560 | UDP-鼠李糖 | 矢车菊色素,矢车菊色素 3-O-葡萄糖苷 | 矢车菊色素 3-O-鼠李糖苷,矢车菊色素 3-O-葡萄糖-鼠李糖苷 | [ |
| 拟南芥 | UGT79B3 | AT4G27570 | UDP-鼠李糖 | 矢车菊色素,矢车菊色素 3-O-葡萄糖苷 | 矢车菊色素 3-O-鼠李糖苷,矢车菊色素 3-O-葡萄糖-鼠李糖苷 | [ |
| 拟南芥 | UGT79B1 | AT5G54060 | UDP-木糖 | 矢车菊色素 3-O-葡萄糖苷 | 矢车菊色素 3-O-木糖(1-2)-葡萄糖苷 | [ |
| 猕猴桃 | F3GT1 | GU079683 | UDP-半乳糖 | 矢车菊色素 | 矢车菊色素 3-O-半乳糖苷 | [ |
| 猕猴桃 | F3GGT1 | FG404013 | UDP-木糖 | 矢车菊色素 3-O-半乳糖苷 | 矢车菊色素 3-O-木糖(1-2)-半乳糖苷 | [ |
| 猕猴桃 | AcUFGT3a | AYJ72756 | UDP-半乳糖 | 矢车菊色素 | 矢车菊色素 3-O-半乳糖苷 | [ |
| 猕猴桃 | AcUFGT6b | MH817045 | UDP-木糖 | 矢车菊色素 3-O-半乳糖苷 | 矢车菊色素 3-O-木糖-半乳糖苷 | [ |
| 桃 | PpUGT78A1 | - | UDP-半乳糖 | 矢车菊色素 | 矢车菊色素-3-半乳糖苷 | [ |
| 桃 | PpUGT78B | - | UDP-葡萄糖 | 矢车菊色素 | 矢车菊色素-3-葡萄糖苷 | [ |
| 桃 | PpUGT78A2 | - | UDP-葡萄糖 | 矢车菊色素 | 矢车菊色素-3-葡萄糖苷 | [ |
| 荔枝 | LcUFGT1 | KR704274 | UDP-葡萄糖 | 矢车菊色素 | 矢车菊色素 3-O-葡萄糖苷 | [ |
| 柿 | DkFGT | BAI40148 | UDP-半乳糖 | 矢车菊色素 | 矢车菊色素 3-O-半乳糖苷 | [ |
| 胎里红枣 | ZjUGT79B1 | - | UDP-鼠李糖 | 矢车菊色素 3-O-葡萄糖苷 | 矢车菊色素 3-O-芸香糖苷 | [ |
| 草莓 | FaGT1 | AAU09442 | UDP-葡萄糖 | 天竺葵色素,矢车菊色素,芍药色素 | 天竺葵色素 3-O-葡萄糖苷,矢车菊色素 3-O-葡萄糖苷,芍药色素 3-O-葡萄糖苷 | [ |
| 芒果 | rMiUFGalT3 | LC474861 | UDP-半乳糖 | 矢车菊色素,飞燕草色素,天竺葵色素,芍药色素,矮牵牛色素 | 矢车菊色素 3-O-半乳糖苷,飞燕草色素 3-O-半乳糖苷,天竺葵色素 3-O-半乳糖苷,芍药色素 3-O-半乳糖苷,矮牵牛色素 3-O-半乳糖苷 | [ |
| 葡萄 | VvGT1 | - | UDP-葡萄糖 | 矢车菊色素 | 矢车菊色素 3-O-葡萄糖苷 | [ |
| 红卷心菜 | UGT78D5 | LC536577.1 | UDP-葡萄糖 | 矢车菊色素 | 矢车菊色素 5-O-葡萄糖苷 | [ |
| 红卷心菜 | UGT79B45 | LC536579.1 | UDP-葡萄糖 | 矢车菊色素 3-O-葡萄糖苷 | 矢车菊色素 3-O-槐糖苷 | [ |
| 红卷心菜 | UGT79B44 | LC536578.1 | UDP-葡萄糖 | 矢车菊色素 3-O-葡萄糖苷 | 矢车菊色素 3-O-槐糖苷 | [ |
| 小苍兰 | Fh3GT1 | ADK75021.1 | UDP-葡萄糖 | 芍药色素,矮牵牛色素,矢车菊色素,飞燕草色素,天竺葵色素,锦葵色素 | 芍药色素 3-O-葡萄糖苷,矮牵牛色素 3-O-葡萄糖苷,矢车菊色素 3-O-葡萄糖苷,飞燕草色素 3-O-葡萄糖苷,天竺葵色素 3-O-葡萄糖苷,锦葵色素 3-O-葡萄糖苷 | [ |
| 小苍兰 | Fh3GT3 | - | UDP-葡萄糖 | 芍药色素,矮牵牛色素,矢车菊色素 | 芍药色素 3-O-葡萄糖苷,矮牵牛色素 3-O-葡萄糖苷,矢车菊色素 3-O-葡萄糖苷 | [ |
| 牵牛花 | 3GGT | AB192315 | UDP-鼠李糖 | 天竺葵色素 3-O-葡萄糖苷,矢车菊色素 3-O-葡萄糖苷,飞燕草色素 3-O-葡萄糖苷 | 天竺葵色素 3-O-芸香糖苷,矢车菊色素 3-O-芸香糖苷,飞燕草色素 3-O-芸香糖苷 | [ |
| 半边莲 | ABRT2 | LC131336 | UDP-鼠李糖 | 飞燕草色素 3-O-葡糖苷,矢车菊色素 3-O-葡萄糖苷,天竺葵色素 3-O-葡萄糖苷, | 飞燕草色素 3-O-芸香糖苷,矢车菊色素 3-O-芸香糖苷,天竺葵色素 3-O-芸香糖苷 | [ |
| 半边莲 | ABRT4 | LC131337 | UDP-鼠李糖 | 飞燕草色素 3-O-葡萄糖苷,矢车菊色素 3-O-葡萄糖苷,天竺葵色素 3 O-葡萄糖苷,锦葵色素 3-O-葡萄糖苷 | 飞燕草色素 3-O-芸香糖苷,矢车菊色素 3-O-芸香糖苷,天竺葵色素 3-O-芸香糖苷,锦葵色素 3-O-芸香糖苷 | [ |
| 紫芹菜 | AgUCGalT1 | - | UDP-半乳糖 | 矢车菊色素 | 矢车菊色素 3-O-半乳糖苷 | [ |
| 紫萝卜 | DcUCGalT1 | KP319022 | UDP-半乳糖 | 矢车菊色素 | 矢车菊色素 3-O-半乳糖苷 | [ |
| 玫瑰 | RhGT1 | AB201048 | UDP-葡萄糖 | 矢车菊色素,矢车菊色素 5-O-葡萄糖苷 | 矢车菊色素 5-O-葡萄糖苷,矢车菊色素 3,5-O-葡萄糖苷 | [ |
| 白杨 | PtUGT78L1 | - | UDP-半乳糖 | 矢车菊色素 | 矢车菊色素 3-O-半乳糖苷 | [ |
| 黑豆 | UGT78K1 | Glyma07g30180 | UDP-葡萄糖 | 矢车菊色素 | 矢车菊色素 3-O-葡萄糖苷 | [ |
| 龙胆草 | Gt5GT7 | AB363839 | UDP-葡萄糖 | 飞燕草素 3-O-葡萄糖苷,矢车菊色素 3-O-葡萄糖苷,天竺葵色素 3-O-葡萄糖苷,锦葵色素 3-O-葡萄糖苷 | 飞燕草素 3,5-O-葡萄糖苷,矢车菊色素 3,5-O-葡萄糖苷,天竺葵色素 3,5-O-葡萄糖苷,锦葵色素 3,5-O-葡萄糖苷 | [ |
| 紫薯 | Ib3GGT | KF056328 | UDP-葡萄糖 | 矢车菊色素 3-O-葡萄糖苷, 芍药色素 3-O-葡萄糖苷 | 矢车菊色素 3-O-槐糖苷,芍药色素 3-O-槐糖苷 | [ |
| 荷兰鸢尾花 | Ih3GT | AB161175 | UDP-葡萄糖 | 飞燕草素,锦葵色素,芍药色素,矢车菊色素,天竺葵色素 | 飞燕草素 3-O-葡萄糖苷,锦葵色素 3-O-葡萄糖苷,芍药色素 3-O-葡萄糖苷,矢车菊色素 3-O-葡萄糖苷,天竺葵色素 3-O-葡萄糖苷 | [ |
| 茶树 | CsUGT78A15 | - | UDP -半乳糖 | 矢车菊色素,飞燕草色素 | 矢车菊色素 3-O-半乳糖苷,飞燕草色素 3-O-半乳糖苷 | [ |
| 物种 Species | 基因 Gene | GenBank登录号 GenBank accession No. | 转化物种 Conversion of species | 转化方法 Transformation methods | 表型 Phenotype | 结论 Conclusion | 参考文献 Reference |
|---|---|---|---|---|---|---|---|
| 红肉猕猴桃 | F3GT1 | GU079683 | 红肉猕猴桃 | RNAi | RNAi后的红肉猕猴桃果实中很少或没有可见的花青素积累 | F3GT1可能是参与红肉猕猴桃品种中花青素积累的关键基因 | [ |
| 红肉猕猴桃 | F3GGT1 | FG404013 | 猕猴桃 | RNAi | RNAi后的红肉猕猴桃果实中花青素的积累水平降低 | F3GGT1可能负责花青素修饰途径的最终产物 | [ |
| 红肉猕猴桃 | AcUFGT3a | AYJ72756 | 红肉猕猴桃和青苹果果实/红肉猕猴桃 | 过表达(瞬转)/基因沉默 | 过表达AcUFGT3a的红肉猕猴桃和青苹果果肉颜色更红/基因沉默的红肉猕猴桃果实中花青素含量,AcUFGT3a的表达量均降低 | AAcUFGT3a可能参与红肉猕猴桃中花青素的积累 | [ |
| 拟南芥 | UGT75C1 | At4g14090 | 拟南芥 | 突变 | UGT75C1突变体植株完全缺乏花青素 5-O-葡萄糖苷 | UGT75C1可能是花青素 5-O-葡萄糖基转移酶 | [ |
| 拟南芥 | UGT79B2 | - | 拟南芥 | 过表达和RNAi,并给与持续光照 | 与野生型植株相比,过表达UGT79B2基因拟南芥植株下胚轴紫色较深,RNAi后拟南芥植株下胚轴紫色较浅 | UGT79B2可能通过积累花青素赋予植物非生物胁迫耐受性 | [ |
| 拟南芥 | UGT79B3 | - | 拟南芥 | 过表达和RNAi,并给与持续光照 | 与野生型植株相比,过表达拟南芥植株下胚轴紫色较深,RNAi后拟南芥植株下胚轴紫色较浅 | UGT79B3可能通过积累花青素赋予植物非生物胁迫耐受性 | [ |
| 荔枝 | LcUFGT1 | KR704274 | 烟草 | 过表达 | 过表达LcUFGT1转基因烟草的花瓣颜色深红,花丝呈粉红色 | LcUFGT1可能与荔枝果皮中的花青素积累有关 | [ |
| 茶树 | CsUGT78A15 | - | 青苹果 | 过表达(瞬转) | 过表达CsUGT78A15的青苹果果肉颜色变红 | CsUGT78A15的上调促进了苹果果皮中花青素 3-O-半乳糖苷的积累 | [ |
| 蝴蝶兰 | PeUFGT3 | EU427548 | 蝴蝶兰 | RNAi | RNAi后蝴蝶兰植株表现出不同程度的花褪色 | PeUFGT3可能在蝴蝶兰花瓣颜色形成中起着关键作用 | [ |
| 玫瑰 | RrGT116 | MK034140 | 拟南芥/玫瑰 | 过表达/基因沉默 | 过表达RrGT116拟南芥植株颜色变深/RrGT116基因沉默后的玫瑰花瓣颜色变浅 | RrGT1可能在玫瑰花青素合成中起着至关重要的作用 | [ |
| 苦荞麦 | FtUFGT8 | - | 拟南芥 | 过表达 | 与野生型植株相比,过表达FtUFGT8的转基因拟南芥中总花色苷的积累增加 | FtUFGT8可能是苦荞芽中花青素形成的候选基因 | [ |
| 苦荞麦 | FtUFGT15 | - | 拟南芥 | 过表达 | 与野生型植株相比,过表达FtUFGT15的转基因拟南芥中总花色苷的积累增加 | FtUFGT15可能是苦荞芽中花青素形成的候选基因 | [ |
| 苦荞麦 | FtUFGT41 | - | 拟南芥 | 过表达 | 与野生型植株相比,过表达 FtUFGT41 的转基因拟南芥中总花色苷的积累 增加 | FtUFGT41可能是苦荞芽中花青素形成的候选基因 | [ |
| 牵牛花 | Ip3GGT | AB192315 | 牵牛花 | 过表达 | 对照组牵牛花花瓣呈红色,转基因牵牛花花瓣呈现淡红色,转基因植株中矢车菊色素 3-O-槐糖苷积累量显著升高高 | 3GGT可将花青素 3-O-葡萄糖苷转化为花青素 3-O-槐糖苷 | [ |
| 半边莲 | ABRT2 | LC131336 | 半边莲 | 过表达 | 对照组半边莲花瓣呈淡紫色,过表达ABRT2的转基因半边莲花瓣呈蓝色 | ABRT2可能是半边莲花呈现蓝色的关键基因 | [ |
| 半边莲 | ABRT4 | LC131337 | 半边莲 | 过表达 | 对照植株花瓣呈淡紫色,过表达ABRT4的转基因半边莲植株花瓣呈 蓝色 | ABRT4可能是半边莲花瓣呈现蓝色的关键基因 | [ |
| 小苍兰 | Fh3GT1 | - | 矮牵牛 | RNAi | 与野生型相比,RNAi矮牵牛植株花的颜色从深红色变为粉红色 | Fh3GT1可能与小苍兰中花青苷的积累有关 | [ |
| 小苍兰 | Fh5GT1 | - | 烟草 | 过表达 | 与野生型植株相比,过表达Fh5GT1的转基因烟草,花瓣颜色更红 | Fh5GT1可能与小苍兰中花青苷的积累有关 | [ |
| 小苍兰 | Fh5GT2 | - | 烟草 | 过表达 | 与野生型植株相比,过表达Fh5GT2的转基因烟草,花瓣颜色更红 | Fh5GT2可能与小苍兰中花青苷的积累有关 | [ |
| 牡丹 | PhUGT78A22 | OM310997 | 牡丹 | 基因沉默 | 与对照组相比,PhUGT78A22基因的沉默改变了牡丹花蕾的颜色 | PhUGT78A22可能与牡丹花瓣的斑点颜色形成有关 | [ |
Table 2 Plant glycosyltransferases related to anthocyanin modification by functional identification in vivo
| 物种 Species | 基因 Gene | GenBank登录号 GenBank accession No. | 转化物种 Conversion of species | 转化方法 Transformation methods | 表型 Phenotype | 结论 Conclusion | 参考文献 Reference |
|---|---|---|---|---|---|---|---|
| 红肉猕猴桃 | F3GT1 | GU079683 | 红肉猕猴桃 | RNAi | RNAi后的红肉猕猴桃果实中很少或没有可见的花青素积累 | F3GT1可能是参与红肉猕猴桃品种中花青素积累的关键基因 | [ |
| 红肉猕猴桃 | F3GGT1 | FG404013 | 猕猴桃 | RNAi | RNAi后的红肉猕猴桃果实中花青素的积累水平降低 | F3GGT1可能负责花青素修饰途径的最终产物 | [ |
| 红肉猕猴桃 | AcUFGT3a | AYJ72756 | 红肉猕猴桃和青苹果果实/红肉猕猴桃 | 过表达(瞬转)/基因沉默 | 过表达AcUFGT3a的红肉猕猴桃和青苹果果肉颜色更红/基因沉默的红肉猕猴桃果实中花青素含量,AcUFGT3a的表达量均降低 | AAcUFGT3a可能参与红肉猕猴桃中花青素的积累 | [ |
| 拟南芥 | UGT75C1 | At4g14090 | 拟南芥 | 突变 | UGT75C1突变体植株完全缺乏花青素 5-O-葡萄糖苷 | UGT75C1可能是花青素 5-O-葡萄糖基转移酶 | [ |
| 拟南芥 | UGT79B2 | - | 拟南芥 | 过表达和RNAi,并给与持续光照 | 与野生型植株相比,过表达UGT79B2基因拟南芥植株下胚轴紫色较深,RNAi后拟南芥植株下胚轴紫色较浅 | UGT79B2可能通过积累花青素赋予植物非生物胁迫耐受性 | [ |
| 拟南芥 | UGT79B3 | - | 拟南芥 | 过表达和RNAi,并给与持续光照 | 与野生型植株相比,过表达拟南芥植株下胚轴紫色较深,RNAi后拟南芥植株下胚轴紫色较浅 | UGT79B3可能通过积累花青素赋予植物非生物胁迫耐受性 | [ |
| 荔枝 | LcUFGT1 | KR704274 | 烟草 | 过表达 | 过表达LcUFGT1转基因烟草的花瓣颜色深红,花丝呈粉红色 | LcUFGT1可能与荔枝果皮中的花青素积累有关 | [ |
| 茶树 | CsUGT78A15 | - | 青苹果 | 过表达(瞬转) | 过表达CsUGT78A15的青苹果果肉颜色变红 | CsUGT78A15的上调促进了苹果果皮中花青素 3-O-半乳糖苷的积累 | [ |
| 蝴蝶兰 | PeUFGT3 | EU427548 | 蝴蝶兰 | RNAi | RNAi后蝴蝶兰植株表现出不同程度的花褪色 | PeUFGT3可能在蝴蝶兰花瓣颜色形成中起着关键作用 | [ |
| 玫瑰 | RrGT116 | MK034140 | 拟南芥/玫瑰 | 过表达/基因沉默 | 过表达RrGT116拟南芥植株颜色变深/RrGT116基因沉默后的玫瑰花瓣颜色变浅 | RrGT1可能在玫瑰花青素合成中起着至关重要的作用 | [ |
| 苦荞麦 | FtUFGT8 | - | 拟南芥 | 过表达 | 与野生型植株相比,过表达FtUFGT8的转基因拟南芥中总花色苷的积累增加 | FtUFGT8可能是苦荞芽中花青素形成的候选基因 | [ |
| 苦荞麦 | FtUFGT15 | - | 拟南芥 | 过表达 | 与野生型植株相比,过表达FtUFGT15的转基因拟南芥中总花色苷的积累增加 | FtUFGT15可能是苦荞芽中花青素形成的候选基因 | [ |
| 苦荞麦 | FtUFGT41 | - | 拟南芥 | 过表达 | 与野生型植株相比,过表达 FtUFGT41 的转基因拟南芥中总花色苷的积累 增加 | FtUFGT41可能是苦荞芽中花青素形成的候选基因 | [ |
| 牵牛花 | Ip3GGT | AB192315 | 牵牛花 | 过表达 | 对照组牵牛花花瓣呈红色,转基因牵牛花花瓣呈现淡红色,转基因植株中矢车菊色素 3-O-槐糖苷积累量显著升高高 | 3GGT可将花青素 3-O-葡萄糖苷转化为花青素 3-O-槐糖苷 | [ |
| 半边莲 | ABRT2 | LC131336 | 半边莲 | 过表达 | 对照组半边莲花瓣呈淡紫色,过表达ABRT2的转基因半边莲花瓣呈蓝色 | ABRT2可能是半边莲花呈现蓝色的关键基因 | [ |
| 半边莲 | ABRT4 | LC131337 | 半边莲 | 过表达 | 对照植株花瓣呈淡紫色,过表达ABRT4的转基因半边莲植株花瓣呈 蓝色 | ABRT4可能是半边莲花瓣呈现蓝色的关键基因 | [ |
| 小苍兰 | Fh3GT1 | - | 矮牵牛 | RNAi | 与野生型相比,RNAi矮牵牛植株花的颜色从深红色变为粉红色 | Fh3GT1可能与小苍兰中花青苷的积累有关 | [ |
| 小苍兰 | Fh5GT1 | - | 烟草 | 过表达 | 与野生型植株相比,过表达Fh5GT1的转基因烟草,花瓣颜色更红 | Fh5GT1可能与小苍兰中花青苷的积累有关 | [ |
| 小苍兰 | Fh5GT2 | - | 烟草 | 过表达 | 与野生型植株相比,过表达Fh5GT2的转基因烟草,花瓣颜色更红 | Fh5GT2可能与小苍兰中花青苷的积累有关 | [ |
| 牡丹 | PhUGT78A22 | OM310997 | 牡丹 | 基因沉默 | 与对照组相比,PhUGT78A22基因的沉默改变了牡丹花蕾的颜色 | PhUGT78A22可能与牡丹花瓣的斑点颜色形成有关 | [ |
| [1] | Deroles S. Anthocyanin biosynthesis in plant cell cultures: a potential source of natural colourants[M]// Anthocyanins. New York: Springer, 2008: 108-167. |
| [2] |
Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology[J]. Plant Physiol, 2001, 126(2): 485-493.
doi: 10.1104/pp.126.2.485 pmid: 11402179 |
| [3] | Dixon RA, Xie DY, Sharma SB. Proanthocyanidins—a final frontier in flavonoid research?[J]. New Phytol, 2005, 165(1): 9-28. |
| [4] |
Tanaka Y, Brugliera F, Chandler S. Recent progress of flower colour modification by biotechnology[J]. Int J Mol Sci, 2009, 10(12): 5350-5369.
doi: 10.3390/ijms10125350 pmid: 20054474 |
| [5] | Andersen ØM JM. Anthocyanins[J]. In: Encyclopedia of Life Sciences. Chichester: John Wiley & Sons, 2010: 1-12. |
| [6] | Plaza M, Pozzo T, Liu JY, et al. Substituent effects on in vitro antioxidizing properties, stability, and solubility in flavonoids[J]. J Agric Food Chem, 2014, 62(15): 3321-3333. |
| [7] | Yonekura-Sakakibara K, Hanada K. An evolutionary view of functional diversity in family 1 glycosyltransferases[J]. Plant J, 2011, 66(1): 182-193. |
| [8] | Sinnott ML. Catalytic mechanism of enzymic glycosyl transfer[J]. Chem Rev, 1990, 90(7): 1171-1202. |
| [9] | Hashimoto K, Tokimatsu T, Kawano S, et al. Comprehensive analysis of glycosyltransferases in eukaryotic genomes for structural and functional characterization of glycans[J]. Carbohydr Res, 2009, 344(7): 881-887. |
| [10] |
Coutinho PM, Deleury E, Davies GJ, et al. An evolving hierarchical family classification for glycosyltransferases[J]. J Mol Biol, 2003, 328(2): 307-317.
pmid: 12691742 |
| [11] | Ross J, Li Y, Lim E, et al. Higher plant glycosyltransferases[J]. Genome Biol, 2001, 2(2): REVIEWS3004. |
| [12] |
Bowles D, Lim EK, Poppenberger B, et al. Glycosyltransferases of lipophilic small molecules[J]. Annu Rev Plant Biol, 2006, 57: 567-597.
pmid: 16669774 |
| [13] |
Vogt T, Jones P. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family[J]. Trends in Plant Science, 2000, 5(9): 380-386.
doi: 10.1016/s1360-1385(00)01720-9 pmid: 10973093 |
| [14] | Noda N. Recent advances in the research and development of blue flowers[J]. Breed Sci, 2018, 68(1): 79-87. |
| [15] |
Song CK, Härtl K, McGraphery K, et al. Attractive but toxic: emerging roles of glycosidically bound volatiles and glycosyltransferases involved in their formation[J]. Mol Plant, 2018, 11(10): 1225-1236.
doi: S1674-2052(18)30273-9 pmid: 30223041 |
| [16] |
Sønderby IE, Geu-Flores F, Halkier BA. Biosynthesis of glucosinolates—gene discovery and beyond[J]. Trends Plant Sci, 2010, 15(5): 283-290.
doi: 10.1016/j.tplants.2010.02.005 pmid: 20303821 |
| [17] |
Sawai S, Saito K. Triterpenoid biosynthesis and engineering in plants[J]. Front Plant Sci, 2011, 2: 25.
doi: 10.3389/fpls.2011.00025 pmid: 22639586 |
| [18] |
Gleadow RM, Møller BL. Cyanogenic glycosides: synthesis, physiology, and phenotypic plasticity[J]. Annu Rev Plant Biol, 2014, 65: 155-185.
doi: 10.1146/annurev-arplant-050213-040027 pmid: 24579992 |
| [19] |
Brazier-Hicks M, Gershater M, Dixon D, et al. Substrate specificity and safener inducibility of the plant UDP-glucose-dependent family 1 glycosyltransferase super-family[J]. Plant Biotechnol J, 2018, 16(1): 337-348.
doi: 10.1111/pbi.12775 pmid: 28640934 |
| [20] |
Dooner HK, Nelson OE. Controlling element-induced alterations in UDPglucose: flavonoid glucosyltransferase, the enzyme specified by the bronze locus in maize[J]. Proc Natl Acad Sci USA, 1977, 74(12): 5623-5627.
pmid: 16592474 |
| [21] | Caputi L, Malnoy M, Goremykin V, et al. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land[J]. Plant J, 2012, 69(6): 1030-1042. |
| [22] | Lim EK, Bowles DJ. A class of plant glycosyltransferases involved in cellular homeostasis[J]. EMBO J, 2004, 23(15): 2915-2922. |
| [23] | Holton TA, Cornish EC. Genetics and biochemistry of anthocyanin biosynthesis[J]. Plant Cell, 1995, 7(7): 1071-1083. |
| [24] |
Welch CR, Wu QL, Simon JE. Recent advances in anthocyanin analysis and characterization[J]. Curr Anal Chem, 2008, 4(2): 75-101.
doi: 10.2174/157341108784587795 pmid: 19946465 |
| [25] | Stintzing FC, Carle R. Functional properties of anthocyanins and betalains in plants, food, and in human nutrition[J]. Trends Food Sci Technol, 2004, 15(1): 19-38. |
| [26] | Koay SS. Establishment of cell suspension culture of Melastoma malabathricum L. for the production of anthocyanin[D]. Universiti Sains Malaysia, 2008. |
| [27] | Wallace TC, Giusti MM. Anthocyanins in cardiovascular disease prevention[M]//Wallace TC, Giusti MM, eds. Anthocyanins in Health and Disease, 2013: 182-215. |
| [28] | Kähkönen MP, Heinonen M. Antioxidant activity of anthocyanins and their aglycons[J]. J Agric Food Chem, 2003, 51(3): 628-633. |
| [29] | Brouillard R. Chemical structure of anthocyanins[M]// Anthocyanins As Food Colors. Amsterdam: Elsevier, 1982: 1-40. |
| [30] | Takeoka G, Dao LT. Anthocyanins. In: Hurst WJ(ed)Methods of Analysis for Functional Foods and Nutraceuticals[M]. CRC Press Inc., Boca Raton, 2002, pp 219-241 |
| [31] | Yoshihara N, Imayama T, Fukuchi-Mizutani M, et al. cDNA cloning and characterization of UDP-glucose: Anthocyanidin 3-O-glucosyltransferase in Iris hollandica[J]. Plant Sci, 2005, 169(3): 496-501. |
| [32] |
Francis FJ, Markakis PC. Food colorants: anthocyanins[J]. Crit Rev Food Sci Nutr, 1989, 28(4): 273-314.
pmid: 2690857 |
| [33] | Saito N, Tatsuzawa F, Miyoshi K, et al. The first isolation of C-glycosylanthocyanin from the flowers of Tricyrtis formosana[J]. Tetrahedron Lett, 2003, 44(36): 6821-6823. |
| [34] | Tatsuzawa F, Saito N, Miyoshi K, et al. Diacylated 8-C-glucosylcyanidin 3-glucoside from the flowers of Tricyrtis formosana[J]. Chem Pharm Bull, 2004, 52(5): 631-633. |
| [35] |
Offen W, Martinez-Fleites C, Yang M, et al. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification[J]. EMBO J, 2006, 25(6): 1396-1405.
doi: 10.1038/sj.emboj.7600970 pmid: 16482224 |
| [36] |
Ford CM, Boss PK, Hoj PB. Cloning and characterization of Vitis vinifera UDP-glucose: flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize Bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo[J]. J Biol Chem, 1998, 273(15): 9224-9233.
doi: 10.1074/jbc.273.15.9224 pmid: 9535914 |
| [37] |
Yamazaki M, Yamagishi E, Gong ZZ, et al. Two flavonoid glucosyltransferases from Petunia hybrida: molecular cloning, biochemical properties and developmentally regulated expression[J]. Plant Mol Biol, 2002, 48(4): 401-411.
pmid: 11905966 |
| [38] | Tohge T, Nishiyama Y, Hirai MY, et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor[J]. Plant J, 2005, 42(2): 218-235. |
| [39] | Morita Y, Hoshino A, Kikuchi Y, et al. Japanese morning glory dusky mutants displaying reddish-brown or purplish-gray flowers are deficient in a novel glycosylation enzyme for anthocyanin biosynthesis, UDP-glucose: anthocyanidin 3-O-glucoside-2’’-O-glucosyltransferase, due to 4-bp insertions in the gene[J]. Plant J, 2005, 42(3): 353-363. |
| [40] | Ogata J, Kanno Y, Itoh Y, et al. Anthocyanin biosynthesis in roses[J]. Nature, 2005, 435(7043): 757-758. |
| [41] |
Griesser M, Hoffmann T, Bellido ML, et al. Redirection of flavonoid biosynthesis through the down-regulation of an anthocyanidin glucosyltransferase in ripening strawberry fruit[J]. Plant Physiol, 2008, 146(4): 1528-1539.
doi: 10.1104/pp.107.114280 pmid: 18258692 |
| [42] |
Nakatsuka T, Sato K, Takahashi H, et al. Cloning and characterization of the UDP-glucose: anthocyanin 5-O-glucosyltransferase gene from blue-flowered gentian[J]. J Exp Bot, 2008, 59(6): 1241-1252.
doi: 10.1093/jxb/ern031 pmid: 18375606 |
| [43] | Kovinich N, Saleem A, Arnason JT, et al. Functional characterization of a UDP-glucose: flavonoid 3-O-glucosyltransferase from the seed coat of black soybean(Glycine max(L.)Merr.)[J]. Phytochemistry, 2010, 71(11/12): 1253-1263. |
| [44] |
Cheng J, Wei GC, Zhou H, et al. Unraveling the mechanism underlying the glycosylation and methylation of anthocyanins in peach[J]. Plant Physiol, 2014, 166(2): 1044-1058.
doi: 10.1104/pp.114.246876 pmid: 25106821 |
| [45] | Li XJ, Zhang JQ, Wu ZC, et al. Functional characterization of a glucosyltransferase gene, LcUFGT1, involved in the formation of cyanidin glucoside in the pericarp of Litchi chinensis[J]. Physiol Plant, 2016, 156(2): 139-149. |
| [46] | Wang H, Wang C, Fan W, et al. UDP-glucose:anthocyanidin 3-O-glucoside-2’’-O-glucosyltransferase catalyzes glycosyl extension of anthocyanins in purple Ipomoea batatas[J]. Journal of Experimental Botany, 2018, 69(22): 5444-5459. |
| [47] | Meng XY, Li YQ, Zhou TT, et al. Functional differentiation of duplicated flavonoid 3- O-glycosyltransferases in the flavonol and anthocyanin biosynthesis of Freesia hybrida[J]. Front Plant Sci, 2019, 10: 1330. |
| [48] | Horiuchi R, Nishizaki Y, Okawa N, et al. Identification of the biosynthetic pathway for anthocyanin triglucoside, the precursor of polyacylated anthocyanin, in red cabbage[J]. J Agric Food Chem, 2020, 68(36): 9750-9758. |
| [49] |
Ikegami A, Akagi T, Potter D, et al. Molecular identification of 1-Cys peroxiredoxin and anthocyanidin/flavonol 3-O-galactosyltransferase from proanthocyanidin-rich young fruits of persimmon(Diospyros kaki Thunb.)[J]. Planta, 2009, 230(4): 841-855.
doi: 10.1007/s00425-009-0989-0 pmid: 19641937 |
| [50] | Montefiori M, Espley RV, Stevenson D, et al. Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit(Actinidia chinensis)[J]. Plant J, 2011, 65(1): 106-118. |
| [51] |
Veljanovski V, Constabel CP. Molecular cloning and biochemical characterization of two UDP-glycosyltransferases from poplar[J]. Phytochemistry, 2013, 91: 148-157.
doi: 10.1016/j.phytochem.2012.12.012 pmid: 23375153 |
| [52] | Xu ZS, Ma J, Wang F, et al. Identification and characterization of DcUCGalT1, a galactosyltransferase responsible for anthocyanin galactosylation in purple carrot(Daucus carota L.) taproots[J]. Sci Rep, 2016, 6: 27356. |
| [53] | Liu YF, Zhou B, Qi YW, et al. Biochemical and functional characterization of AcUFGT3a, a galactosyltransferase involved in anthocyanin biosynthesis in the red-fleshed kiwifruit(Actinidia chinensis)[J]. Physiol Plant, 2018, 162(4): 409-426. |
| [54] |
Feng K, Xu ZS, Liu JX, et al. Isolation, purification, and characterization of AgUCGalT1, a galactosyltransferase involved in anthocyanin galactosylation in purple celery(Apium graveolens L.)[J]. Planta, 2018, 247(6): 1363-1375.
doi: 10.1007/s00425-018-2870-5 pmid: 29520459 |
| [55] | Katayama-Ikegami A, Byun Z, Okada S, et al. Characterization of the Recombinant UDP: flavonoid 3-O-galactosyltransferase from Mangifera indica ‘Irwin’(MiUFGalT3)involved in Skin Coloring[J]. Hortic J, 2020, 89(5): 516-524. |
| [56] |
Kanzaki S, Kamikawa S, Ichihi A, et al. Isolation of UDP: Flavonoid 3-O-glycosyltransferase (UFGT)-like genes and expression analysis of genes associated with anthocyanin accumulation in mango ‘Irwin’ skin[J]. Horticulture Journal, 2019, 88(4): 435-443.
doi: 10.2503/hortj.UTD-098 |
| [57] | Li P, Li YJ, Zhang FJ, et al. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation[J]. Plant J, 2017, 89(1): 85-103. |
| [58] | Hsu YH, Tagami T, Matsunaga K, et al. Functional characterization of UDP-rhamnose-dependent rhamnosyltransferase involved in anthocyanin modification, a key enzyme determining blue coloration in Lobelia erinus[J]. Plant J, 2017, 89(2): 325-337. |
| [59] | Yonekura-Sakakibara K, Fukushima A, Nakabayashi R, et al. Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana[J]. Plant J, 2012, 69(1): 154-167. |
| [60] | Liu YF, Liu J, Qi YW, et al. Identification and characterization of AcUFGT6b, a xylosyltransferase involved in anthocyanin modification in red-fleshed kiwifruit(Actinidia chinensis)[J]. Plant Cell Tissue Organ Cult PCTOC, 2019, 138(2): 257-271. |
| [61] | Shi QQ, Du JT, Zhu DJ, et al. Metabolomic and transcriptomic analyses of anthocyanin biosynthesis mechanisms in the color mutant Ziziphus jujuba cv. tailihong[J]. J Agric Food Chem, 2020, 68(51): 15186-15198. |
| [62] | He XQ, Huang RH, Liu LP, et al. CsUGT78A15 catalyzes the anthocyanidin 3-O-galactoside biosynthesis in tea plants[J]. Plant Physiol Biochem, 2021, 166: 738-749. |
| [63] | Sui X, Gao X, Ao M, et al. cDNA cloning and characterization of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in Freesia hybrida[J]. Plant Cell Rep, 2011, 30(7): 1209-1218. |
| [64] |
Yao PF, Deng RY, Huang YJ, et al. Diverse biological effects of glycosyltransferase genes from Tartary buckwheat[J]. BMC Plant Biol, 2019, 19(1): 339.
doi: 10.1186/s12870-019-1955-z pmid: 31382883 |
| [65] | Ju ZG, Sun W, Meng XY, et al. Isolation and functional characterization of two 5-O-glucosyltransferases related to anthocyanin biosynthesis from Freesia hybrida[J]. Plant Cell Tissue Organ Cult PCTOC, 2018, 135(1): 99-110. |
| [66] | Sun C, Wang CM, Zhang W, et al. The R2R3-type MYB transcription factor MdMYB90-like is responsible for the enhanced skin color of an apple bud sport mutant[J]. Hortic Res, 2021, 8(1): 156. |
| [67] | Qian MJ, Sun YW, Allan AC, et al. The red sport of ‘Zaosu’ pear and its red-striped pigmentation pattern are associated with demethylation of the PyMYB10 promoter[J]. Phytochemistry, 2014, 107: 16-23. |
| [68] | Matus JT, Cavallini E, Loyola R, et al. A group of grapevine MYBA transcription factors located in chromosome 14 control anthocyanin synthesis in vegetative organs with different specificities compared with the berry color locus[J]. Plant J, 2017, 91(2): 220-236. |
| [69] | Chen WH, Hsu CY, Cheng HY, et al. Downregulation of putative UDP-glucose: flavonoid 3-O-glucosyltransferase gene alters flower coloring in Phalaenopsis[J]. Plant Cell Rep, 2011, 30(6): 1007-1017. |
| [70] | Sui XM, Zhao MY, Han X, et al. RrGT1, a key gene associated with anthocyanin biosynthesis, was isolated from Rosa rugosa and identified via overexpression and VIGS[J]. Plant Physiol Biochem, 2019, 135: 19-29. |
| [71] | Li Y, Kong F, Liu Z, Peng L, et al. PhUGT78A22, a novel glycosyltransferase in Paeonia ‘He Xie’, can catalyze the transfer of glucose to glucosylated anthocyanins during petal blotch formation[J]. BMC Plant Biol. 2022, 22(1):1-15. |
| [72] | An XH, Tian Y, Chen KQ, et al. The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation[J]. J Plant Physiol, 2012, 169(7): 710-717. |
| [73] | Liu XF, Yin XR, Allan AC, et al. The role of MrbHLH1 and MrMYB1 in regulating anthocyanin biosynthetic genes in tobacco and Chinese bayberry(Myrica rubra)during anthocyanin biosynthesis[J]. Plant Cell Tissue Organ Cult, 2013, 115(3): 285-298. |
| [74] | Xu HF, Wang N, Liu JX, et al. The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes[J]. Plant Mol Biol, 2017, 94(1/2): 149-165. |
| [75] | Grützner R, König K, Horn C, et al. A transient expression tool box for anthocyanin biosynthesis in Nicotiana benthamiana[J]. Plant Biotechnol J, 2024, 22(5): 1238-1250. |
| [1] | YANG Jia-hong, LI Jing-yi, WU Jia-hao, HUANG You-mei, LIU Yan-fen, QIN Yuan, CAI Han-yang. Research Progress in the Auxin Signaling Pathway Involved in the Regulation of Female Gametophyte Development in Arabidopsis [J]. Biotechnology Bulletin, 2024, 40(7): 19-27. |
| [2] | LONG Jing, CHEN Jing-min, LIU Xiao, ZHANG Yi-fan, ZHOU Li-bin, DU Yan. Repair Mechanisms of DNA Double-strand Breaks and Their Roles in Heavy Ion Mutagenesis and Gene Editing in Plants [J]. Biotechnology Bulletin, 2024, 40(7): 55-67. |
| [3] | YUAN Hai-peng YE Yun-shu SI Hao JI Qiu-yan ZHANG Yu-hong. Effects of Arbuscular Mycorrhizal Fungi on Plant Stress Resistance and Secondary Metabolite Synthesis [J]. Biotechnology Bulletin, 2024, 40(6): 45-56. |
| [4] | WANG Yu-shu, ZHAO Lin-lin, ZHAO Shuang, HU Qi, BAI Hui-xia, WANG Huan, CAO Ye-ping, FAN Zhen-yu. Cloning and Expression Analysis of BrCYP83B1 Gene in Chinese Cabbage [J]. Biotechnology Bulletin, 2024, 40(6): 152-160. |
| [5] | SUN Ya-nan, WANG Chun-xue, WANG Xin, DU Bing-hai, LIU Kai, WANG Cheng-qiang. Biocontrol Characteristics of Bacillus atrophaeus CNY01 and Its Salt-resistant and Growth-promoting Effect on Maize Seedling [J]. Biotechnology Bulletin, 2024, 40(5): 248-260. |
| [6] | SONG Hui, CAO Wen-gang, XIAO Xiao-wen, DU Jun. Development and Application of Antibody Modified DNA Sequencing Enzymes [J]. Biotechnology Bulletin, 2024, 40(5): 84-93. |
| [7] | WANG Li-chao, LI Huan, SHENG Ruo-cheng, LI Min, CHEN Feng-mao. Role of Acetylation in the Pathogenic Process of Plant Pathogens [J]. Biotechnology Bulletin, 2024, 40(5): 1-12. |
| [8] | WU Wei, MA Qiu-gang, ZHU Xuan, WANG Jian. Research Progress in the High-value Utilization of Lignocellulose Biomass by Steam Explosion [J]. Biotechnology Bulletin, 2024, 40(5): 23-37. |
| [9] | GUO Hui-yan, DONG Xue, AN Meng-nan, XIA Zi-hao, WU Yuan-hua. Research Progress in the Functions of Key Enzymes of Ubiquitination Modification in Plant Stress Responses [J]. Biotechnology Bulletin, 2024, 40(4): 1-11. |
| [10] | GAO Zhi-wei, WEI Ming, YU Zu-long, WU Guo-qiang, WEI Jun-long. Identification of Salt-tolerant Plant Growth-promoting Bacterium W-1 and Its Effect on the Salt-tolerance of Sainfoin(Onobrychis viciaefolia) [J]. Biotechnology Bulletin, 2024, 40(4): 217-227. |
| [11] | PENG Yu-jia, LI Wen-cui, LIU Yong-bo. Research Progress in the Evolution Mechanisms for Insect Resistance to Insecticides and Bt-transgenic Plants [J]. Biotechnology Bulletin, 2024, 40(4): 40-51. |
| [12] | WANG Juan, WANG Xin, TIAN Qin, MA Xiao-mei, ZHOU Xiao-feng, LI Bao-cheng, DONG Cheng-guang. Association Analysis and Exploration of Elite Alleles of Plant Architecture Traits in Gossypium hirsutum L. [J]. Biotechnology Bulletin, 2024, 40(3): 146-154. |
| [13] | FANG Tian-yi, YUE Yan-ling. Mechanisms of Root Exudates-mediated Plant Resistance to Soil-borne Diseases [J]. Biotechnology Bulletin, 2024, 40(3): 52-61. |
| [14] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [15] | YUAN Xin-yu, ZHONG Cai-hong, ZHANG Long, ZHENG Hao, LI Ji-tao, ZHANG Qiong. Identification and Function Analysis of AcMYB88 in Kiwifruit (Actinidia chinensis) [J]. Biotechnology Bulletin, 2024, 40(2): 183-196. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||