








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (1): 208-217.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0511
Previous Articles Next Articles
ZENG Ting1,2,3( ), ZHANG Lan1,2, LUO Rui1(
), ZHANG Lan1,2, LUO Rui1( )
)
Received:2025-05-18
Online:2026-01-26
Published:2026-02-04
Contact:
LUO Rui
E-mail:1774837790@qq.com;rluo1@gzu.edu.cn
ZENG Ting, ZHANG Lan, LUO Rui. Functional Analysis of the Transcription Factor MpR2R3-MYB17 in Regulating Gemma Development in Marchantia polymorpha L.[J]. Biotechnology Bulletin, 2026, 42(1): 208-217.
| 引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) | 用途 Usage |
|---|---|---|
| M-F | GCTCTAGAATGGGGAGAGCGCCTTG | 基因克隆 |
| M-R | GCCCCCGGGTCAGCGCTCCATGACGGC | |
| gRT1 | TGCTGCGATAAGCAGGGGTTgttttagagctagaaat | sgRNA表达盒构建 |
| gRT2 | CGGGCACTTCCAAAACATGCgttttagagctagaaat | |
| gRNA-R | CGGAGGAAAATTCCATCCAC | |
| P-F | AGCGTGggtctcGCTCGACGCGTATCCATCCACTCCAAGCTC | |
| P-R | TTCAGAggtctcTACCGACTAGTATGGAATCGGCAGCAAAGG | |
| GUS-F | TCTGCGACGCTCACACCGAT | 过表达阳性植株鉴定 |
| GUS-R | GCCAACGCGCAATATGCCTT | |
| qMpR2R3-MYB17-F | CTACTGGAACACGCACCTGA | 实时荧光定量PCR |
| qMpR2R3-MYB17-R | AGATGACTCAACTTGGAGCACA | |
| qMpAPT-F | CGAAAGCCCAAGAAGCTACC | 内参基因 |
| qMpAPT-R | GTACCCCCGGTTGCAATAAG | |
| Hyg-F | GTCGTCCATCACAGTTTGCC | 突变体阳性鉴定 |
| Hyg-R | TGCTTGACATTGGGGAGTTT | |
| PegMYB17-F | TATATTTGCCGTTGCTGCCG | 测序片段克隆 |
| PegMYB17-R | AGGGTTCAAAGCGCGATTCA |
Table 1 Primers used in this study
| 引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) | 用途 Usage |
|---|---|---|
| M-F | GCTCTAGAATGGGGAGAGCGCCTTG | 基因克隆 |
| M-R | GCCCCCGGGTCAGCGCTCCATGACGGC | |
| gRT1 | TGCTGCGATAAGCAGGGGTTgttttagagctagaaat | sgRNA表达盒构建 |
| gRT2 | CGGGCACTTCCAAAACATGCgttttagagctagaaat | |
| gRNA-R | CGGAGGAAAATTCCATCCAC | |
| P-F | AGCGTGggtctcGCTCGACGCGTATCCATCCACTCCAAGCTC | |
| P-R | TTCAGAggtctcTACCGACTAGTATGGAATCGGCAGCAAAGG | |
| GUS-F | TCTGCGACGCTCACACCGAT | 过表达阳性植株鉴定 |
| GUS-R | GCCAACGCGCAATATGCCTT | |
| qMpR2R3-MYB17-F | CTACTGGAACACGCACCTGA | 实时荧光定量PCR |
| qMpR2R3-MYB17-R | AGATGACTCAACTTGGAGCACA | |
| qMpAPT-F | CGAAAGCCCAAGAAGCTACC | 内参基因 |
| qMpAPT-R | GTACCCCCGGTTGCAATAAG | |
| Hyg-F | GTCGTCCATCACAGTTTGCC | 突变体阳性鉴定 |
| Hyg-R | TGCTTGACATTGGGGAGTTT | |
| PegMYB17-F | TATATTTGCCGTTGCTGCCG | 测序片段克隆 |
| PegMYB17-R | AGGGTTCAAAGCGCGATTCA |
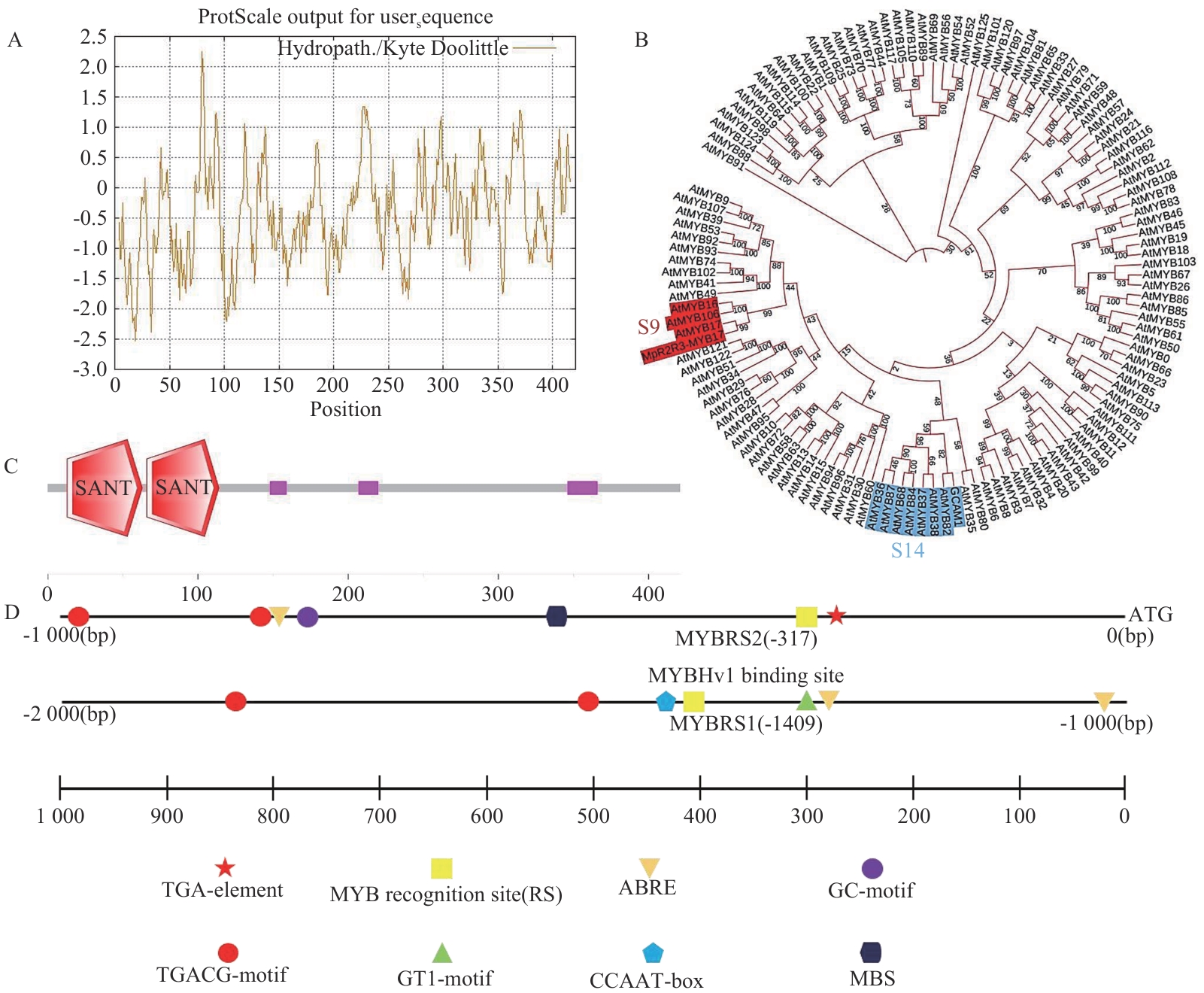
Fig. 1 Bioinformatics analysis and phylogenetic treeA: Hydrophilicity/hydrophobicity prediction of theMpR2R3-MYB17 protein amino acid sequence. B: Phylogenetic tree. C: Conserved domain analysis of the MpR2R3-MYB17 protein (Red: SANT domain. Purple: Low-complexity region). D: Analysis of cis-regulatory elements in the MpR2R3-MYB17 gene promoter region
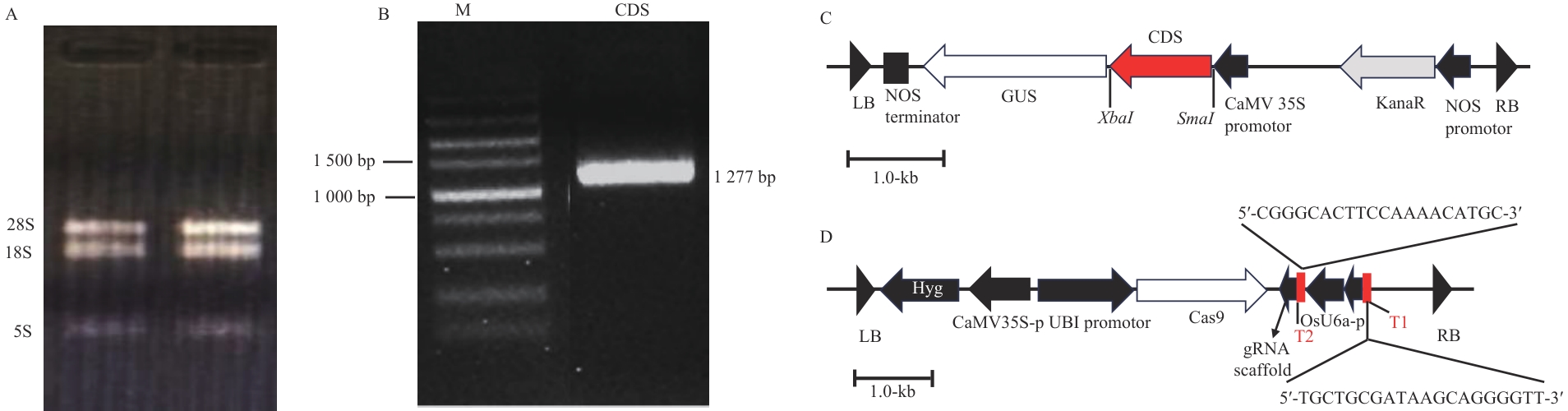
Fig. 2 RNA extraction, cloning of MpR2R3-MYB17 gene, and recombinant plasmid mapsA: RNA extraction. B: MpR2R3-MYB17 gene cloning, the product length is 1 277 bp. C: pBI121-MpR2R3-MYB17 recombinant plasmid map (labelled gene). D: pEGCas9Pubi-MpR2R3-MYB17 recombinant plasmid (T1 and T2: sgRNA target sequences. gRNA scaffold: gRNA structural backbone)

Fig. 4 Screening of resistant plants, PCR identification of overexpressed plants and GUS stainingA: Marchantia polymorpha gemmae following Agrobacterium tumefaciens infection. B: Kanamycin-resistant plants selected after transformation. C: PCR confirmation of MpR2R3-MYB17-overexpressed transgenic lines. D: Histochemical GUS staining in MpR2R3-MYB17-overexpressed plants
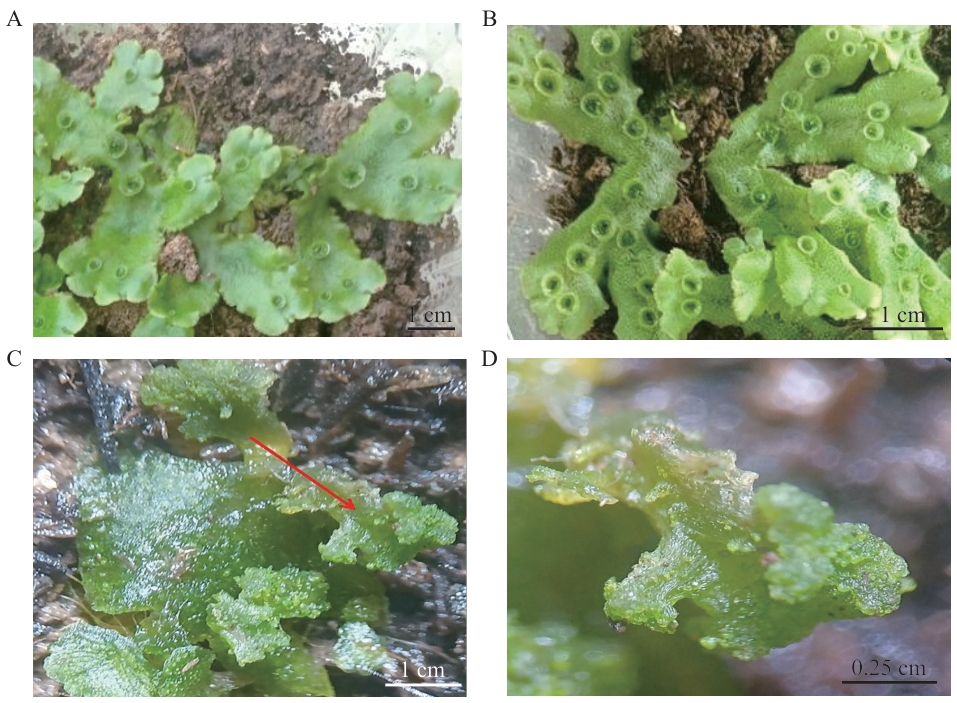
Fig. 5 Phenotype of overexpressing plantsA: Wild type of M. polymorpha L.. B: Phenotypes of overexpressing plant lines (increased gemma cup density). C: Phenotypes of overexpressing plant lines (undifferentiated cell clumps). Undifferentiated cell clumps are indicated by red arrows. D: Enlarged view of undifferentiated cell clumps from panel C
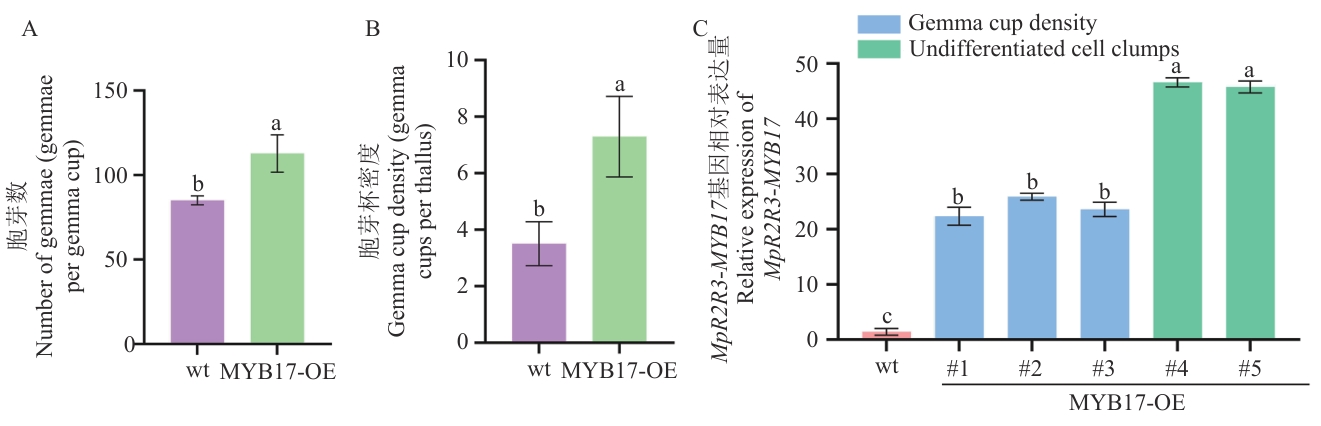
Fig. 6 Phenotypic statistics and expression analysis of overexpressed plant linesA: Number of gemmae per gemma cup (gemmae/gemma cup). B: Gemma cup density (gemma cups per thallus). C: Relative expression of MpR2R3-MYB17 (wt: wild-type. MYB17-OE#1/2/3: Overexpressed lines with increased gemma cup density. MYB17-OE#4/5: Overexpressed lines producing undifferentiated cell clumps). Different lowercase letters indicate significant differences between groups (P<0.05)
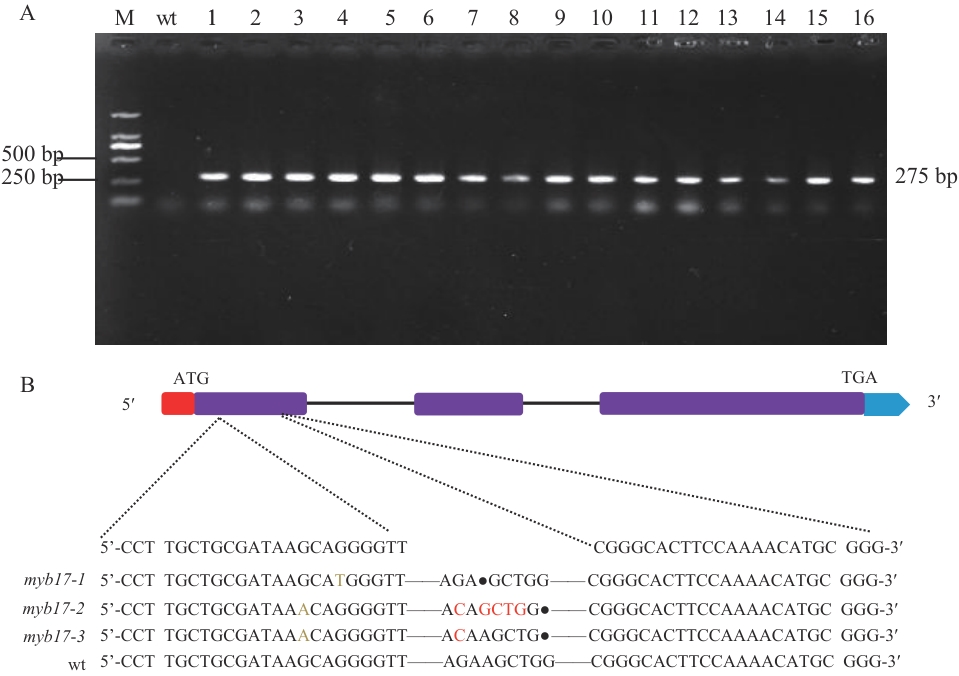
Fig. 7 Mutant identification and sequence alignmentA: PCR identification of transgenic plants. B: Mutant sequence alignment (red box: 5′ non-coding region; purple boxes: exons; blue box: 3′ non-coding region. In the sequence alignment, red letters denote nucleotide substitutions. Dots "·": Base deletions. Horizontal line "—": Unmutated bases that are consistent with the reference sequence and are omitted)
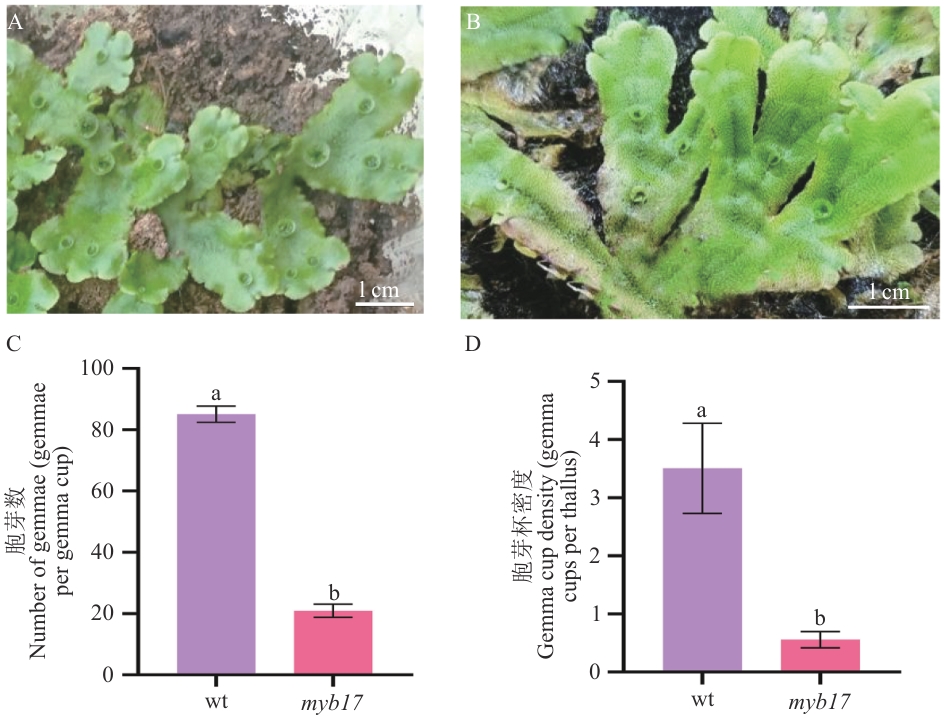
Fig. 8 Phenotypic analysis of mutantsA: Wild type of M. polymorpha L.. B:Mutant lines. C:Number of gemmae per gemma cup (gemmae/gemma cup). D: Gemma cup density (gemma cups per thallus). myb17 indicates the phenotypic statistics of three independent mutant lines, myb17-1, myb17-2 and myb17-3, and different lowercase letters indicate significant differences between groups (P<0.01)
| [1] | Beaulieu C, Libourel C, Mbadinga Zamar DL, et al. The Marchantia polymorpha pangenome reveals ancient mechanisms of plant adaptation to the environment [J]. Nat Genet, 2025, 57(3): 729-740. |
| [2] | Shimamura M. Marchantia polymorpha: taxonomy, phylogeny and morphology of a model system [J]. Plant Cell Physiol, 2016, 57(2): 230-256. |
| [3] | Kato H, Yasui Y, Ishizaki K. Gemma cup and gemma development in Marchantia polymorpha [J]. New Phytol, 2020, 228(2): 459-465. |
| [4] | 胡雅丹, 伍国强, 刘晨, 等. MYB转录因子在调控植物响应逆境胁迫中的作用 [J]. 生物技术通报, 2024, 40(6): 5-22. |
| Hu YD, Wu GQ, Liu C, et al. Roles of MYB transcription factor in regulating the responses of plants to stress [J]. Biotechnol Bull, 2024, 40(6): 5-22. | |
| [5] | Zhang DW, Zhou HP, Zhang Y, et al. Diverse roles of MYB transcription factors in plants [J]. J Integr Plant Biol, 2025, 67(3): 539-562. |
| [6] | Lal M, Bhardwaj E, Chahar N, et al. Comprehensive analysis of 1R-and 2R-MYBs reveals novel genic and protein features, complex organisation, selective expansion and insights into evolutionary tendencies [J]. Funct Integr Genomics, 2022, 22(3): 371-405. |
| [7] | Yang JH, Zhang BH, Gu G, et al. Genome-wide identification and expression analysis of the R2R3-MYB gene family in tobacco (Nicotiana tabacum L.) [J]. BMC Genomics, 2022, 23(1): 432. |
| [8] | Koo Y, Poethig RS. Expression pattern analysis of three R2R3-MYB transcription factors for the production of anthocyanin in different vegetative stages of Arabidopsis leaves [J]. Appl Biol Chem, 2021, 64(1): 5. |
| [9] | Qin W, Xie LH, Li YP, et al. An R2R3-MYB transcription factor positively regulates the glandular secretory trichome initiation in Artemisia annua L [J]. Front Plant Sci, 2021, 12: 657156. |
| [10] | Okumura T, Nomoto Y, Kobayashi K, et al. MYB3R-mediated active repression of cell cycle and growth under salt stress in Arabidopsis thaliana [J]. J Plant Res, 2021, 134(2): 261-277. |
| [11] | Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis [J]. Trends Plant Sci, 2010, 15(10): 573-581. |
| [12] | Preston J, Wheeler J, Heazlewood J, et al. AtMYB32 is required for normal pollen development in Arabidopsis thaliana [J]. Plant J, 2004, 40(6): 979-995. |
| [13] | Steiner-Lange S, Unte US, Eckstein L, et al. Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers [J]. Plant J, 2003, 34(4): 519-528. |
| [14] | Yasui Y, Tsukamoto S, Sugaya T, et al. GEMMA CUP-ASSOCIATED MYB1, an ortholog of axillary meristem regulators, is essential in vegetative reproduction in Marchantia polymorpha [J]. Curr Biol, 2019, 29(23): 3987-3995.e5. |
| [15] | Sugano SS, Nishihama R. CRISPR/Cas9-based genome editing of transcription factor genes in Marchantia polymorpha [J]. Methods Mol Biol, 2018, 1830: 109-126. |
| [16] | Du YM, Liu YF, Hu JX, et al. CRISPR/Cas9 systems: Delivery technologies and biomedical applications [J]. Asian J Pharm Sci, 2023, 18(6): 100854. |
| [17] | Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res, 2002, 30(1): 325-327. |
| [18] | Chen ZW, Halford NG, Liu CH. Real-time quantitative PCR primer design, reference gene selection, calculations and statistics [J]. Metabolites, 2023, 13(7): 806. |
| [19] | Maren NA, Duduit JR, Huang DB, et al. Stepwise optimization of real-time RT-PCR analysis [J]. Methods Mol Biol, 2023, 2653: 317-332. |
| [20] | Naramoto S, Hata Y, Fujita T, et al. The bryophytes Physcomitrium patens and Marchantia polymorpha as model systems for studying evolutionary cell and developmental biology in plants [J]. Plant Cell, 2022, 34(1): 228-246. |
| [21] | 关范圆, 郑语嫣, 米芯雨, 等. R2R3型MYB转录因子调控植物胁迫应答的研究进展 [J]. 植物医学, 2024, 3(3): 1-9. |
| Guan FY, Zheng YY, Mi XY, et al. Research advance of the functions of R2R3-MYB transcription factors in plant response to biotic and abiotic stresses [J]. Plant Health Med, 2024, 3(3): 1-9. | |
| [22] | 田琴, 刘奎, 吴翔纬, 等. 转录因子VcMYB17调控蓝莓抗旱性的功能研究 [J]. 生物技术通报, 2025, 41(4): 198-210. |
| Tian Q, Liu K, Wu XW, et al. Functional study of transcription factor VcMYB17 in regulating drought tolerance in blueberry [J]. Biotechnol Bull, 2025, 41(4): 198-210. | |
| [23] | Brockington SF, Alvarez-Fernandez R, Landis JB, et al. Evolutionary analysis of the MIXTA gene family highlights potential targets for the study of cellular differentiation [J]. Mol Biol Evol, 2013, 30(3): 526-540. |
| [24] | Jakoby MJ, Falkenhan D, Mader MT, et al. Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106 [J]. Plant Physiol, 2008, 148(3): 1583-1602. |
| [25] | 张哲, 何鑫玺, 张纪利, 等. NtMYB17过表达对烟草腺毛发育的影响 [J]. 中国烟草科学, 2024, 45(3): 86-92. |
| Zhang Z, He XX, Zhang JL, et al. Effect of NtMYB17 overexpression on development of glandular trichomes in tobacco [J]. Chin Tob Sci, 2024, 45(3): 86-92. | |
| [26] | Machado A, Wu YR, Yang YM, et al. The MYB transcription factor GhMYB25 regulates early fibre and trichome development [J]. Plant J, 2009, 59(1): 52-62. |
| [27] | Tak H, Negi S, Ganapathi TR. Overexpression of MusaMYB31, a R2R3 type MYB transcription factor gene indicate its role as a negative regulator of lignin biosynthesis in banana [J]. PLoS One, 2017, 12(2): e0172695. |
| [28] | Proust H, Honkanen S, Jones VAS, et al. RSL class I genes controlled the development of epidermal structures in the common ancestor of land plants [J]. Curr Biol, 2016, 26(1): 93-99. |
| [29] | Flores-Sandoval E, Eklund DM, Hong SF, et al. Class C ARFs evolved before the origin of land plants and antagonize differentiation and developmental transitions in Marchantia polymorpha [J]. New Phytol, 2018, 218(4): 1612-1630. |
| [30] | Aki SS, Mikami T, Naramoto S, et al. Cytokinin signaling is essential for organ formation in Marchantia polymorpha [J]. Plant Cell Physiol, 2019, 60(8): 1842-1854. |
| [1] | LONG Lin-xi, ZENG Yin-ping, WANG Qian, DENG Yu-ping, GE Min-qian, CHEN Yan-zhuo, LI Xin-juan, YANG Jun, ZOU Jian. Identification of Sunflower GH3 Gene Family and Analysis of Their Function in Flower Development [J]. Biotechnology Bulletin, 2026, 42(1): 125-138. |
| [2] | REN Yun-er, WU Guo-qiang, CHENG Bin, WEI Ming. Genome-wide Identification of the BvATGs Genes Family in Sugar Beet (Beta vulgaris L.) and Analysis of Their Expression Pattern under Salt Stress [J]. Biotechnology Bulletin, 2026, 42(1): 184-197. |
| [3] | YANG Juan, FENG Hui, JI Nai-zhe, SUN Li-ping, WANG Yun, ZHANG Jia-nan, ZHAO Shi-wei. Cloning and Functional Analysis of AP2/ERF Transcription Factors RcERF4 and RcRAP2-12 in Rose [J]. Biotechnology Bulletin, 2026, 42(1): 150-160. |
| [4] | ZHANG Chi-hao, LIU Jin-nan, CHAO Yue-hui. Cloning and Functional Analysis of a bZIP Transcription Factor MtbZIP29 from Medicago truncatula [J]. Biotechnology Bulletin, 2026, 42(1): 241-250. |
| [5] | WU Cui-cui, CHEN Deng-ke, LAN Gang, XIA Zhi, LI Peng-bo. Bioinformatics Analysis of Peanut Transcription Factor AhHDZ70 and Its Tolerances to Salt and Drought [J]. Biotechnology Bulletin, 2026, 42(1): 198-207. |
| [6] | WANG Bin, LIN Chong, YUAN Xiao, JIANG Yuan-yuan, WANG Yu-kun, XIAO Yan-hui. Cloning of bHLH Transcription Factor UNE10 and Its Regulatory Roles in the Biosynthesis of Volatile Compounds in Clove Basil [J]. Biotechnology Bulletin, 2025, 41(9): 207-218. |
| [7] | CHEN Qiang, YU Ying-fei, ZHANG Ying, ZHANG Chong. Regulatory Effect of Methyl Jasmonate on Postharvest Chilling Injury in Oriental Melon ‘Emerald’ [J]. Biotechnology Bulletin, 2025, 41(9): 105-114. |
| [8] | XU Xiao-ping, YANG Cheng-long, HE Xing, GUO Wen-jie, WU Jian, FANG Shao-zhong. Cloning of the LoAPS1 and Its Function Analysis during the Process of Dormancy Release in Lilium [J]. Biotechnology Bulletin, 2025, 41(9): 195-206. |
| [9] | DONG Xiang-xiang, MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song. Bioinformatics Analysis and Flowering Regulation Function of FveBBX20 Gene in Woodland Strawberry [J]. Biotechnology Bulletin, 2025, 41(9): 115-123. |
| [10] | HUANG Shi-yu, TIAN Shan-shan, YANG Tian-wei, GAO Man-rong, ZHANG Shang-wen. Genome-wide Identification and Expression Pattern Analysis of WRI1 Gene Family in Erythropalum scandens [J]. Biotechnology Bulletin, 2025, 41(8): 242-254. |
| [11] | LAI Shi-yu, LIANG Qiao-lan, WEI Lie-xin, NIU Er-bo, CHEN Ying-e, ZHOU Xin, YANG Si-zheng, WANG Bo. The Role of NbJAZ3 in the Infection of Nicotiana benthamiana by Alfalfa Mosaic Virus [J]. Biotechnology Bulletin, 2025, 41(8): 186-196. |
| [12] | LI Ya-qiong, GESANG La-mao, CHEN Qi-di, YANG Yu-huan, HE Hua-zhuan, ZHAO Yao-fei. Heterologous Overexpression of Sorghum SbSnRK2.1 Enhances the Resistance to Salt Stress in Arabidopsis [J]. Biotechnology Bulletin, 2025, 41(8): 115-123. |
| [13] | ZENG Dan, HUANG Yuan, WANG Jian, ZHANG Yan, LIU Qing-xia, GU Rong-hui, SUN Qing-wen, CHEN Hong-yu. Genome-wide Identification and Expression Analysis of bZIP Transcription Factor Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2025, 41(8): 197-210. |
| [14] | YU Yong-xia, DU Zai-hui, ZHU Long-jiao, XU Wen-tao. Application and Research Progress of Gene Editing Technology in Bovine [J]. Biotechnology Bulletin, 2025, 41(8): 34-41. |
| [15] | LI Kai-jie, WU Yao, LI Dan-dan. Cloning of Gene CtbHLH128 in Safflower and Response Function Regulating Drought Stress [J]. Biotechnology Bulletin, 2025, 41(8): 234-241. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||