








生物技术通报 ›› 2021, Vol. 37 ›› Issue (1): 272-281.doi: 10.13560/j.cnki.biotech.bull.1985.2020-0389
李信申1( ), 黄小梅2, 吴淑秀3, 黄瑞荣1, 魏林根4, 华菊玲1(
), 黄小梅2, 吴淑秀3, 黄瑞荣1, 魏林根4, 华菊玲1( )
)
收稿日期:2020-04-04
出版日期:2021-01-26
发布日期:2021-01-15
作者简介:李信申,男,博士,研究方向:植物细菌病害防控;E-mail: 基金资助:
LI Xin-shen1( ), HUANG Xiao-mei2, WU Shu-xiu3, HUANG Rui-rong1, WEI Lin-gen4, HUA Ju-ling1(
), HUANG Xiao-mei2, WU Shu-xiu3, HUANG Rui-rong1, WEI Lin-gen4, HUA Ju-ling1( )
)
Received:2020-04-04
Published:2021-01-26
Online:2021-01-15
摘要:
为实现植物青枯病的早期诊断,需要建立一种适于田间快速便捷检测青枯病菌的方法。以细胞色素C基因为靶标设计一套特异性引物,建立了植物青枯病的LAMP检测方法。此方法最低检测极限为1 pg,可在1 h 内完成,不依赖昂贵复杂的仪器,结果可经肉眼观察。利用此方法,在人工接种发病的茄子、番茄、花生、芝麻和凹头苋茎部浸出液和马铃薯病薯块茎组织液中均检测出青枯病菌的存在,尤其适用于田间疑似罹病的芝麻、花生、番茄、马铃薯和甘薯等植株的检测,且LAMP法的检出率远高于PCR法。应用LAMP技术检测青枯病菌快速高效、特异性强、灵敏度高,操作简单,适于在基层推广运用。
李信申, 黄小梅, 吴淑秀, 黄瑞荣, 魏林根, 华菊玲. 植物青枯病菌环介导等温扩增快速检测技术研究[J]. 生物技术通报, 2021, 37(1): 272-281.
LI Xin-shen, HUANG Xiao-mei, WU Shu-xiu, HUANG Rui-rong, WEI Lin-gen, HUA Ju-ling. Rapid Detection of Plant Bacterial Wilt by Loop-mediated Isothermal Amplification[J]. Biotechnology Bulletin, 2021, 37(1): 272-281.
| 菌株编号 | 菌株名称 | 寄主 | 演化型/生理小种 |
|---|---|---|---|
| 1 | 青枯病菌菌株Ejxnc01 | 茄子 | Ⅰ/1 |
| 2 | 青枯病菌菌株Cjxnc01 | 辣椒 | Ⅰ/1 |
| 3 | 青枯病菌菌株Tjxjx01 | 番茄 | Ⅰ/1 |
| 4 | 青枯病菌菌株Pjxga01 | 花生 | Ⅰ/1 |
| 5 | 青枯病菌菌株Seppx05 | 芝麻 | Ⅰ/1 |
| 6 | 青枯病菌菌株JSja03 | 凹头苋 | Ⅰ/1 |
| 7 | 青枯病菌菌株Tjxga03 | 烟草 | Ⅰ/1 |
| 8 | 青枯病菌菌株Gjxjj01 | 姜 | Ⅰ/4 |
| 9 | 青枯病菌菌株Pjxnc01 | 马铃薯 | Ⅱ/3 |
| 10 | 青枯病菌菌株Pjxga02 | 马铃薯 | Ⅱ/3 |
| 11 | 青枯病菌菌株Pjxjj03 | 马铃薯 | Ⅱ/3 |
| 12 | 青枯病菌菌株Pjxda04 | 马铃薯 | Ⅱ/3 |
| 13 | 蒲桃雷尔氏菌R001 | 丁香树 | - |
| 14 | 皮氏伯克霍尔德氏菌Bp001 | 土壤 | - |
表1 用于检测引物特异性的青枯病菌和非青枯病菌菌株
| 菌株编号 | 菌株名称 | 寄主 | 演化型/生理小种 |
|---|---|---|---|
| 1 | 青枯病菌菌株Ejxnc01 | 茄子 | Ⅰ/1 |
| 2 | 青枯病菌菌株Cjxnc01 | 辣椒 | Ⅰ/1 |
| 3 | 青枯病菌菌株Tjxjx01 | 番茄 | Ⅰ/1 |
| 4 | 青枯病菌菌株Pjxga01 | 花生 | Ⅰ/1 |
| 5 | 青枯病菌菌株Seppx05 | 芝麻 | Ⅰ/1 |
| 6 | 青枯病菌菌株JSja03 | 凹头苋 | Ⅰ/1 |
| 7 | 青枯病菌菌株Tjxga03 | 烟草 | Ⅰ/1 |
| 8 | 青枯病菌菌株Gjxjj01 | 姜 | Ⅰ/4 |
| 9 | 青枯病菌菌株Pjxnc01 | 马铃薯 | Ⅱ/3 |
| 10 | 青枯病菌菌株Pjxga02 | 马铃薯 | Ⅱ/3 |
| 11 | 青枯病菌菌株Pjxjj03 | 马铃薯 | Ⅱ/3 |
| 12 | 青枯病菌菌株Pjxda04 | 马铃薯 | Ⅱ/3 |
| 13 | 蒲桃雷尔氏菌R001 | 丁香树 | - |
| 14 | 皮氏伯克霍尔德氏菌Bp001 | 土壤 | - |
| 引物名称 | 引物序列(5'-3') | 长度/bp |
|---|---|---|
| F3 | AGCGGTGCCAATCCGTA | 17 |
| B3 | TGCCATGGTCAGGTACTGAT | 20 |
| FIP | AGCAATCCGAAGGTGCCGAATGTCGCGTA- CAACCAGGA | 38 |
| BIP | TCGGTATCCCGACAACACCATGTGGGCGT- CGATCGCATA | 39 |
表2 用于环式扩增细胞色素C信号肽的引物序列
| 引物名称 | 引物序列(5'-3') | 长度/bp |
|---|---|---|
| F3 | AGCGGTGCCAATCCGTA | 17 |
| B3 | TGCCATGGTCAGGTACTGAT | 20 |
| FIP | AGCAATCCGAAGGTGCCGAATGTCGCGTA- CAACCAGGA | 38 |
| BIP | TCGGTATCCCGACAACACCATGTGGGCGT- CGATCGCATA | 39 |
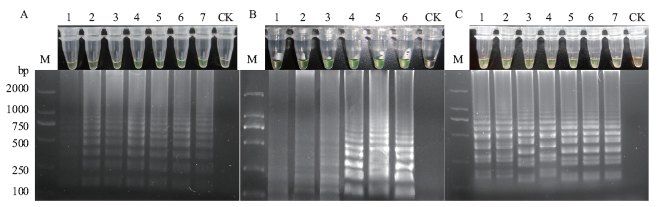
图1 LAMP反应体系的优化 (A)M:Marker DL2000;1:0 mmol/L;2:2 mmol/L;3:4 mmol/L;4:6 mmol/L;5:8 mmol/L;6:10 mmol/L;7:12 mmol/L;CK:阴性对照;(B)M:Marker DL2000;1-6.内引物浓度/外引物浓度分别为2∶1,4∶1,6∶1,8∶1,10∶1,12∶1;CK:阴性对照;(C)M:Marker DL2000;1:59℃;2:60℃;3:61℃;4:62℃;5:63℃;6:64℃;7:65℃;CK:阴性对照
| 来源植物 | 取样部位 | 样本数量 | LAMP检测 | PCR检测 | ||||
|---|---|---|---|---|---|---|---|---|
| 检出总数 | 检出率 | 检出总数 | 检出率 | |||||
| 芝麻 | 茎杆 | 25 | 25 | 100% | 23 | 92.00% | ||
| 花生 | 茎杆 | 24 | 23 | 95.83% | 21 | 87.50% | ||
| 番茄 | 叶片 | 23 | 23 | 100% | 21 | 91.30% | ||
| 马铃薯 | 块茎 | 24 | 23 | 95.83% | 22 | 91.67% | ||
| 甘薯 | 块茎 | 22 | 21 | 95.45% | 20 | 90.91% | ||
表3 疑似罹病植株样本LAMP和PCR检测结果
| 来源植物 | 取样部位 | 样本数量 | LAMP检测 | PCR检测 | ||||
|---|---|---|---|---|---|---|---|---|
| 检出总数 | 检出率 | 检出总数 | 检出率 | |||||
| 芝麻 | 茎杆 | 25 | 25 | 100% | 23 | 92.00% | ||
| 花生 | 茎杆 | 24 | 23 | 95.83% | 21 | 87.50% | ||
| 番茄 | 叶片 | 23 | 23 | 100% | 21 | 91.30% | ||
| 马铃薯 | 块茎 | 24 | 23 | 95.83% | 22 | 91.67% | ||
| 甘薯 | 块茎 | 22 | 21 | 95.45% | 20 | 90.91% | ||
| [1] |
Mansfield J, Genin S, Magori S, et al. Top 10 plant pathogenic bacteria in molecular plant pathology[J]. Molecular Plant Pathology, 2012,13(6):614-629.
doi: 10.1111/j.1364-3703.2012.00804.x URL pmid: 22672649 |
| [2] | Patil VU, Girimalla V, Sagar V, et al. Genome sequencing of four strains of Phylotype I, II and IV of Ralstonia solanacearum that cause potato bacterial wilt in India[J]. Brazilian Journal of Microbiology, 2017,48(2):193-195. |
| [3] | Bocsanczy AM, Espindola AS, Norman DJ. Whole-genome sequences of Ralstonia solanacearum strains P816, P822, and P824, emerging pathogens of blueberry in Florida[J]. Microbiology Resource Announcements, 2019,8(3):e01316-18. |
| [4] |
Jiang G, Wei Z, Xu J, et al. Bacterial wilt in China:history, current status, and future perspectives[J]. Frontiers in Plant Science, 2017,8:1549.
URL pmid: 28955350 |
| [5] | Álvarez B, López1 MM, Biosca EG. Biocontrol of the major plant pathogen Ralstonia solanacearum in irrigation water and host plants by novel waterborne lytic bacteriophages[J]. Fronter in Microbiology, 2019,10:2813. |
| [6] | Tan X, Qiu H, Li F, et al. Complete genome sequence of sequevar 14M Ralstonia solanacearum strain HA4-1 reveals novel type III effectors acquired through horizontal gene transfer[J]. Fronter in Microbiology, 2019,10:1893. |
| [7] | Wang H, Qi J, Xiao D, et al. A re-evaluation of dilution for eliminating PCR inhibition in soil DNA samples[J]. Soil Biology and Biochemistry, 2017,106:109-118. |
| [8] | Gorny AM, Wang X, Hay FS, et al. Development of a species-specific PCR for detection and quantification of Meloidogyne hapla in soil using the 16D10 root-knot nematode effector gene[J]. Plant Disease, 2019,103(8):1902-1909. |
| [9] | Villa JE, Tsuchiya K, Horita M, et al. Phylogenetic relationships of Ralstonia solanacearum species complex strains from Asia and other continents based on 16S rDNA, endoglucanase, and hrpB gene sequences[J]. Journal of General Plant Pathology, 2005,71(1):39-46. |
| [10] | Singh D, Sinha S, Chaudhary G, et al. Biological characterization and genetic diversity of Indian strains of Ralstonia solanacearum Biovars 3 and 4 causing bacterial wilt of tomato[J]. Journal of Plant Pathology and Microbiology, 2018,9(443):2. |
| [11] |
Caruso P, Bertolini E, Cambra M, et al. A new and sensitive co-operational polymerase chain reaction for rapid detection of Ralstonia solanacearum in water[J]. Journal of Microbiological Methods, 2003,55(1):257-272.
URL pmid: 14500017 |
| [12] | Vreeburg RAM, Zendman AJW, Pol A, et al. Validation of four real-time TaqMan PCR s for the detection of Ralstonia solanacearum and/or Ralstonia pseudosolanacearum and/or Clavibacter michiganensis subsp. sepedonicus in potato tubers using a statistical regression approach[J]. EPPO Bulletin, 2018,48(1):86-96. |
| [13] | Kubota R, Vine BG, Alvarez AM, Jenkins DM. Detection of Ralstonia solanacearum by loop-mediated isothermal amplification[J]. Bacteriology, 2008,98(9):1045-1051. |
| [14] | Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA[J]. Nucleic Acids Research, 2000,28(12):e63. |
| [15] | 贾蒙骜, 陈兴江, 林叶春, 等. 基于环式等温扩增的烟草青枯病病原菌快速检测方法[J]. 中国农业大学学报, 2014,19(1):93-98. |
| Jia MA, Chen XJ, Lin YC, et al. Rapid and sensitive detection method for Ralstonia solanacearum based on Loop-mediated isothermal amplification[J]. Journal of China Agricultural University, 2014,19(1):93-98. | |
| [16] | Morisset D, Pirc M, Llop P, et al. Loop-mediated isothermal amplification of specific endoglucanase gene sequence for detection of the bacterial wilt pathogen Ralstonia solanacearum[J]. PLoS One, 2014,9(4):e96027. |
| [17] | 黄雯, 徐进, 张昊, 等. 植物青枯菌LAMP检测方法的建立[J]. 中国农业科学, 2016,49(11):2093-2102. |
| Huang W, Xu J, Zhang H, et al. Development of a LAMP approach for detection of Ralstonia solanacearum[J]. Scientia Agricultura Sinica, 2016,49(11):2093-2102. | |
| [18] | Okiro LA, Tancos MA, Nyanjom SG, et al. Comparative evaluation of LAMP, qPCR, conventional PCR, and ELISA to detect Ralstonia solanacearum in Kenyan potato fields[J]. Plant Disease, 2019,103(5):959-965. |
| [19] | Notomi T, Mori Y, Tomita N, et al. Loop-mediated isothermal amplification(LAMP):principle, features, and future prospects[J]. Journal of Microbiology, 2015,53(1):1-5. |
| [20] | 华菊玲, 胡白石, 李湘民, 等. 芝麻细菌性青枯病病原菌及其生化变种鉴定[J]. 植物保护学报, 2012,39(1):39-44. |
| Hua JL, Hu BS, Li XM, et al. Identification of the pathogen causing bacterial wilt of sesame and its biovars[J]. Acta Phytophylacica Sinica, 2012,39(1):39-44. | |
| [21] |
El Sayed T, Samuel J, Nour E, et al. Biocontrol of bacterial wilt disease through complex interaction between tomato plant, antagonists, the indigenous rhizosphere microbiota and Ralstonia solanacearum[J]. Frontiers in Microbiology, 2019,10:2835.
doi: 10.3389/fmicb.2019.02835 URL pmid: 31998244 |
| [22] | Rajapaksha R, Kohombange S, Dissanayake D, et al. Bitter gourd(Memordica charantia L.), new host of Ralstonia solanacearum in Sri Lanka[J]. International Journal of Recent Innovations in Academic Research, 2019,3(7):111-118. |
| [23] |
Lin CH, Chuang MH, Wang JF. First report of bacterial wilt caused by Ralstonia solanacearum on Chard in Taiwan[J]. Plant Disease, 2015,99(2):282-282.
doi: 10.1094/PDIS-07-14-0780-PDN URL pmid: 30699590 |
| [24] | Jiang Y, Li B, Liu P, et al. First report of bacterial wilt caused by Ralstonia solanacearum on fig trees in China[J]. Forest Pathology, 2016,46(3):256-258. |
| [25] | Weibel J, Tran TM, Bocsanczy AM, et al. A Ralstonia solanacearum strain from Guatemala infects diverse flower crops, including new asymptomatic hosts vinca and sutera, and causes symptoms in geranium, mandevilla vine, and new host African daisy(Osteospermum ecklonis)[J]. Plant Health Progress, 2016,17(2):114-121. |
| [26] | Huang Q, Allen C. Polygalacturonases are required for rapid colonization and full virulence of Ralstonia solanacearum on tomato plants[J]. Physiological and Molecular Plant Pathology, 2000,57(2):77-83. |
| [27] | Caldwell D, Kim BS, Iyer-Pascuzzi AS. Ralstonia solanacearum differentially colonizes roots of resistant and susceptible tomato plants[J]. Phytopathology, 2017,107(5):528-536. |
| [28] | Weibel J, Tran TM, Bocsanczy AM, et al. A Ralstonia solanacearum strain from Guatemala infects diverse flower crops, including new asymptomatic hosts vinca and sutera, and causes symptoms in geranium, mandevilla vine, and new host African daisy(Osteospe-rmum ecklonis)[J]. Plant Health Progress, 2016,17(2):114-121. |
| [29] | Sagcan H, Kara NT. Detection of Potato ring rot Pathogen Clavibacter michiganensis subsp. s epedonicus by Loop-mediated isothermal amplification(LAMP)assay[J]. Scientific Reports, 2019,9(1):1-8. |
| [30] | Chen ZD, Kang HJ, Chai AL, et al. Development of a loop-mediated isothermal amplification(LAMP)assay for rapid detection of Pseudomonas syringae pv. tomato in planta[J]. European Journal of Plant Pathology, 2020,156:739-750. |
| [31] | Ocenar J, Arizala D, Boluk G, et al. Development of a robust, field-deployable loop-mediated isothermal amplification(LAMP)assay for specific detection of potato pathogen Dickeya dianthicola targeting a unique genomic region[J]. PLoS One, 2019,14(6):e0218868. |
| [32] | Dobhal S, Larrea-Sarmiento A, Alvarez AM, et al. Development of a loop-mediated isothermal amplification assay for specific detection of all known subspecies of Clavibacter michiganensis[J]. Journal of Applied Microbiology, 2019,126(2):388-401. |
| [33] | Drais MI, Maheshwari Y, Selvaraj V, et al. Development and validation of a loop-mediated isothermal amplification technique(LAMP)for the detection of Spiroplasma citri, the causal agent of citrus stubborn disease[J]. European Journal of Plant Pathology, 2019,155(1):125-134. |
| [34] | 陈瑞朋. 烟草野火病菌与角斑病菌LAMP快速检测方法的建立与应用[D]. 泰安:山东农业大学, 2018. |
| Chen RP. Development and application of a loop-mediated isothermal amplification method for rapid detection of Pseudomonas syringae pv. tabaci and Pseudomonas syringae pv. Angulata[D]. Taian:Shandong Agricultural University, 2018. | |
| [35] | Sun M, Liu H, Huang J, et al. A loop-mediated isothermal amplification assay for rapid detection of Pectobacterium aroidearum that causes soft rot in Konjac[J]. International Journal of Molecular Sciences, 2019,20(8):1937. |
| [36] | Ghosh DK, Bhose S, BhoWarghanese A, et al. Loop-mediated isothermal amplification(LAMP)based method for rapid and sensitive detection of ‘Candidatus Liberibacter asiaticus’ in citrus and the psyllid vector, Diaphorina citri Kuwayama[J]. Journal of Plant Biochemistry and Biotechnology, 2016,25(2):219-223. |
| [37] | Kalpage HA, Bazylianska V, Recanati MA, et al. Tissue-specific regulation of cytochrome c by post-translational modifications:respiration, the mitochondrial membrane potential, ROS, and apoptosis[J]. The FASEB Journal, 2019,33(2):1540-1553. |
| [38] | Kang MJ, Lee MH, Shim JK, et al. PCR-based specific detection of Ralstonia solanacearum by amplification of cytochrome c1 signal peptide sequences[J]. Journal of Microbiology and Biotechnology, 2007,17(11):1765-1771. |
| [39] | 胡利伟, 牟文君, 郭建华, 等. 基于锁核苷酸(LNA)增敏的植烟土壤青枯雷尔氏菌定量PCR检测[J]. 烟草科技, 2017,50(12):14-21. |
| Hu LW, Mu WJ, Guo JH, et al. Locked nucleic acid-enhanced quantitative real-time PCR detection of Ralstonia solanacearum in tobacco planting soil[J]. Tobacco Science and Technology, 2017,50(12):14-21. | |
| [40] | Lau YL, Lai MY, Teoh BT, et al. Colorimetric detection of dengue by single tube reverse-transcription-loop-mediated isothermal amplification[J]. PLoS One, 2015,10(9):e0138694. |
| [41] | Salinas NR, Little DP. Electric LAMP:virtual loop-mediated isothermal amplification[J]. ISRN Bioinform, 2012,21:696758. |
| [42] | Santiago-Felipe S, Tortajada-Genaro LA, Carrascosa J, et al. Real-time loop-mediated isothermal DNA amplification in compact disc micro-reactors[J]. Biosensors and Bioelectronics, 2016,79:300-306. |
| [43] | Wong YP, Othman S, Lau YL, et al. Loop-mediated isothermal amplification(LAMP):a versatile technique for detection of micro-organisms[J]. Journal of Applied Microbiology, 2018,124(3):626-643. |
| [44] | 应淑敏, 郭俭, 王教瑜, 等. 环介导等温扩增技术在植物病原物检测中的应用[J]. 植物保护学报, 2020,47(2):234-244. |
| Ying SM, Guo J, Wang JY, et al. Application of LAMP in the detection of plant pathogens[J]. Acta Phytophylacica Sinica, 2020,47(2):234-244. | |
| [45] | Chen HW, Weissenberger G, Ching WM. Development of lyophilized loop-mediated isothermal amplification reagents for the detection of Leptospira[J]. Military Medicine, 2016,181(S5):227-231. |
| [46] | Huang M, Zhou X, Wang H, et al. Clustered regularly interspaced short palindromic repeats/Cas9 triggered isothermal amplification for site-specific nucleic acid detection[J]. Analytical Chemistry, 2018,90(3):2193-2200. |
| [47] | Teng F, Guo L, Cui T, et al. CDetection:CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity[J]. Genome Biology, 2019,20(1):1-7. |
| [48] | Bao Y, Jiang Y, Xiong E, et al. CUT-LAMP:Contamination-free loop-mediated isothermal amplification based on the CRISPR/Cas9 cleavage[J]. ACS Sensors, 2020,5(4):1082-1091. |
| [49] | 林晟豪, 杜再慧, 张秀杰, 等. 基于环介导等温扩增技术的生物传感器研究进展[J]. 生物技术进展, 2019,9(6):599-610. |
| Lin SH, Du ZH, Zhang XJ, et al. Progress on biosensors based on loop-mediated isothermal amplification[J]. Current Biotechnology, 2019,9(6):599-610. | |
| [50] | 高威芳, 章礼平, 朱鹏. 等温扩增技术及其结合CRISPR在微生物快速检测中的研究进展[J]. 生物技术通报, 2020,36(5):22-31. |
| Gao WF, Zhang LP, Zhu P. Recent progress on isothermal amplification technology and its combination with CRISPR in rapid detection of microorganisms[J]. Biotechnology Bulletin, 2020,36(5):22-31. | |
| [51] | Mukama O, de Dieu Habimana J, Meng X, et al. Synergetic performance of isothermal amplification techniques and lateral flow approach for nucleic acid diagnostics[J]. Analytical Biochemistry, 2020: 113762. |
| [1] | 陈晓琳, 刘洋儿, 许文涛, 郭明璋, 刘慧琳. 合成生物学细胞传感技术在食品安全快速检测中的应用[J]. 生物技术通报, 2023, 39(1): 137-149. |
| [2] | 李佳乐, 林晟豪, 许文涛. 基于环介导等温扩增的抗虫基因超灵敏比色生物传感器构建[J]. 生物技术通报, 2022, 38(8): 69-76. |
| [3] | 付志强, 熊艳. 便携式生物光学传感器的研究进展[J]. 生物技术通报, 2021, 37(3): 219-226. |
| [4] | 赵颖, 王楠, 陆安祥, 冯晓元, 郭晓军, 栾云霞. 核酸适配体侧流层析分析技术在真菌毒素检测中的应用[J]. 生物技术通报, 2020, 36(8): 217-227. |
| [5] | 高威芳, 章礼平, 朱鹏. 等温扩增技术及其结合CRISPR在微生物快速检测中的研究进展[J]. 生物技术通报, 2020, 36(5): 22-31. |
| [6] | 王琦, 颜春蕾, 高洪伟, 吴薇, 杨庆利. 基于核酸适配体传感器检测食品致病菌的研究进展[J]. 生物技术通报, 2020, 36(11): 245-258. |
| [7] | 李秋芬, 刘启臣, 徐爱玲, 张艳, 宋志文. 基于LAMP的好氧反硝化菌筛选及其反硝化性能[J]. 生物技术通报, 2019, 35(9): 210-217. |
| [8] | 林惠娇, 杨华卫, 古恒森, 蒋湘, 张海磊, 刘昱辰, 周而勋. 三种重要水果褐腐病菌快速检测试纸的研制[J]. 生物技术通报, 2019, 35(6): 205-212. |
| [9] | 郭沛, 赵龙, 胡翮. 公共场所水环境中活嗜肺军团菌的快速检测方法[J]. 生物技术通报, 2019, 35(3): 203-209. |
| [10] | 李子微, 邓仲良. 土拉弗朗西斯氏菌LAMP快速检测方法的应用[J]. 生物技术通报, 2019, 35(2): 212-217. |
| [11] | 张微, 李志新, 付春江, 刘卫平. 马铃薯S病毒胶体金免疫层析试纸条的研制[J]. 生物技术通报, 2019, 35(12): 184-188. |
| [12] | 梁玉林, 刘秀 ,周鹏飞 ,周振森 ,尹建军. 基于反转录环介导等温扩增技术检测大肠杆菌O157[J]. 生物技术通报, 2018, 34(6): 59-65. |
| [13] | 袁向芬, 吕继洲, 吴绍强, 王巧黎, 赵宏. 环介导等温扩增技术作为基因快速诊断方法的研究与展望[J]. 生物技术通报, 2018, 34(10): 64-70. |
| [14] | 王大洲,郭天笑,郑实,商颖,许文涛. 核酸等温扩增技术在微生物快速检测中的研究进展[J]. 生物技术通报, 2017, 33(7): 49-61. |
| [15] | 谢文萍, 肇慧君, 张琳, 姜红旭, 吴斌, 孙浩. LAMP技术快速诊断布鲁氏菌病的研究[J]. 生物技术通报, 2017, 33(3): 186-192. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||