








生物技术通报 ›› 2022, Vol. 38 ›› Issue (2): 195-204.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0389
收稿日期:2021-03-29
出版日期:2022-02-26
发布日期:2022-03-09
作者简介:李毅丹,男,博士,副研究员,研究方向:玉米赤霉素代谢调控;E-mail: 基金资助:Received:2021-03-29
Published:2022-02-26
Online:2022-03-09
摘要:
赤霉素作为重要的植物激素,参与了植物诸多发育过程的调控。一些涉及赤霉素生物合成和信号传导途径的重要调控基因对作物的株型、产量和品质能够产生积极的影响,已在农业生产中得到广泛应用。其中,Rht-1和sd-1等位基因由于分别赋予了小麦和水稻半矮化的特性,从而促成了20世纪后半叶的“绿色革命”。本文回顾了与“绿色革命”相关的赤霉素代谢调控在影响作物株高、产量、养分利用等方面的研究成果,并对未来如何合理开发和利用赤霉素调控基因,培育出更多的“绿色革命”作物品种进行了展望。
李毅丹, 单晓辉. 赤霉素代谢调控与绿色革命[J]. 生物技术通报, 2022, 38(2): 195-204.
LI Yi-dan, SHAN Xiao-hui. Gibberellin Metabolism Regulation and Green Revolution[J]. Biotechnology Bulletin, 2022, 38(2): 195-204.
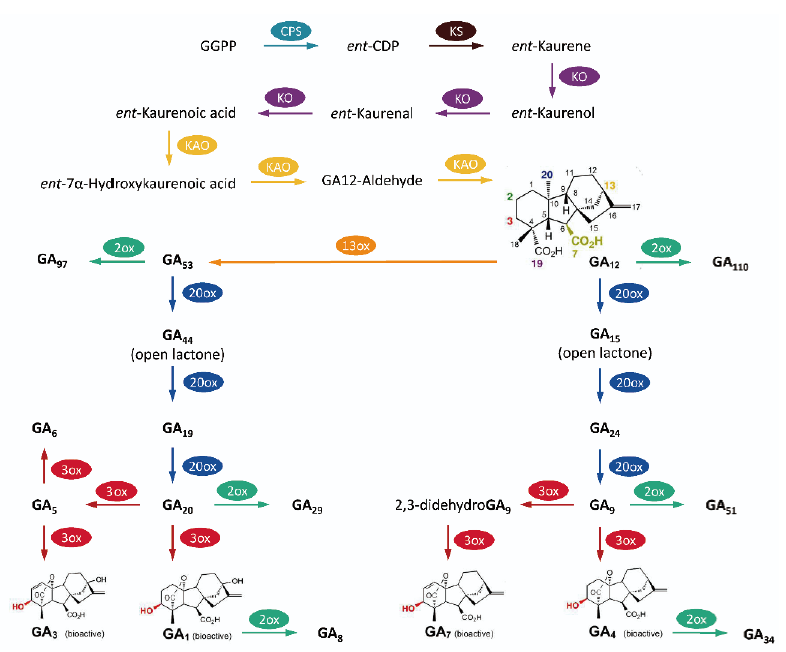
图1 植物中赤霉素的生物合成途径示意图 2ox:GA 2-氧化酶;3ox:GA 3-氧化酶;13ox:GA 13-氧化酶;20ox:GA 20-氧化酶;GGPP:牻牛儿基牻牛儿基焦磷酸;CPS:ent-copalyl二磷酸合酶;KS:ent-kaurene合酶;KO:ent-Kaurene氧化酶;KAO:ent-kaurenoic酸氧化酶。此图修改自Yamaguchi[2]
Fig. 1 Gibberellin biosynthesis pathways in plants 2ox:GA 2-oxidase. 3ox:GA 3-oxidase. 13ox:GA 13-oxidase. 20ox:GA 20-oxidase. GGDP:geranylgeranyl diphosphate. CPS:ent-copalyl diphosphate synthase. KS:ent-kaurene synthase. KO:ent-kaurene oxidase. KAO:ent-kaurenoic acid oxidase. This figure was modified from Yamaguchi[2]

图2 GA-GID1-DELLA信号传导途径示意图 GID1与生物活性的GA结合会导致其构象发生变化,从而促进与DELLA蛋白的相互作用。F-box蛋白的募集启动了SCF E3泛素连接酶介导的DELLA的泛素化,泛素化后的DELLA会进一步的被蛋白酶降解。DELLA的缺失减轻了其对生长抑制并抑制了其他DELLA介导的反应
Fig. 2 Sketch of GA-GID1-DELLAsignal transduction Binding with a bioactive GA results in a conformational change in the GID1 receptor,thus which promotes interaction with DELLA proteins. Recruitment of an F-box protein initiates ubiquitination of DELLA mediated by an SCF E3 ubiquitin ligase targeting the DELLA for proteasomal degradation. Deficiency of DELLA eliminates its growth repression and suppresses other DELLA-mediated reactions
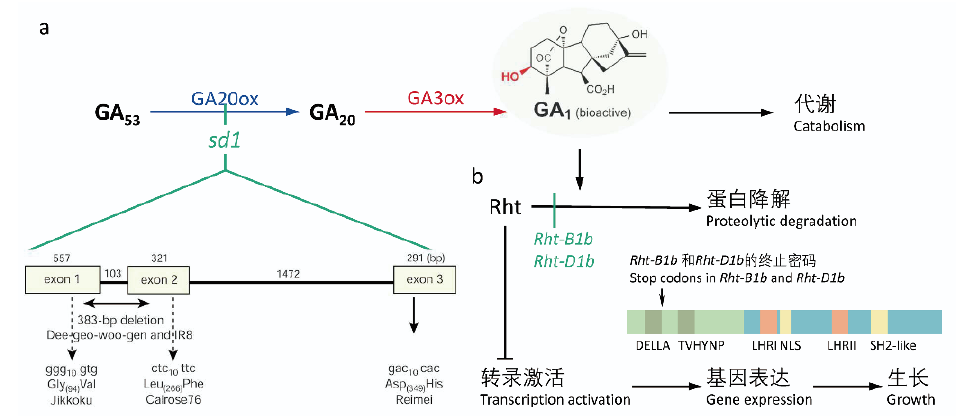
图3 赤霉素合成和信号传导途径中的“绿色革命”基因 a:sd1等位基因的突变位点。sd1突变影响了GA20ox的活性。该酶负责催化GA53转化为GA20,生成生物活性产物GA1。GA20ox由3个外显子和2个内含子组成。每个突变分别用箭头(替换)或横线(缺失)来表示;b:GA是可导致生长抑制蛋白Rht的降解。突变型的Rht(Rht-B1b和Rht-D1b)对GAs诱导的降解不敏感。箭头表示Rht-B1b和Rht-D1b中终止密码的位置。C末端(浅蓝色)在所有相关蛋白中高度保守,并含有抑制因子活性。N端(绿色)是GAs信号识别域,包括2个GAs诱导降解所必需的高度保守的序列(深蓝色)。此图修改自Hedden等[6]和Sasaki等[9]3 水稻“绿色革命”基因semidwarf-1(sd-1)
Fig.3 Green Revolution genes in gibberellin biosynthesis and signal transduction a:Mutation sites of the sd1 alleles. sd1 mutations affect the activity of GA20ox,which catalyzes the conversion of GA53 to GA20 in the biosynthetic pathway to the biological active product,GA1. The GA20ox gene consists of three exons and two introns. The mutation in each allele is indicated by either an arrow(single-nucleotide substitution)or a line(internal deletion). b:GA action results in the degradation of Rht. The mutant forms of Rht(Rht-B1b and Rht-D1b)are not susceptible to GA-induced degradation. The arrow indicates the position of terminal codons in Rht-B1b and Rht-D1b. The C-terminus(light blue)is highly conserved in all related proteins and contains the repressor activity. The N-terminus(green)contains the GA-signalling domain that includes two highly conserved motifs(dark blue)that are required for GA-induced degradation. This figure was modified from Hedden et al[6]and Sasaki et al[9]
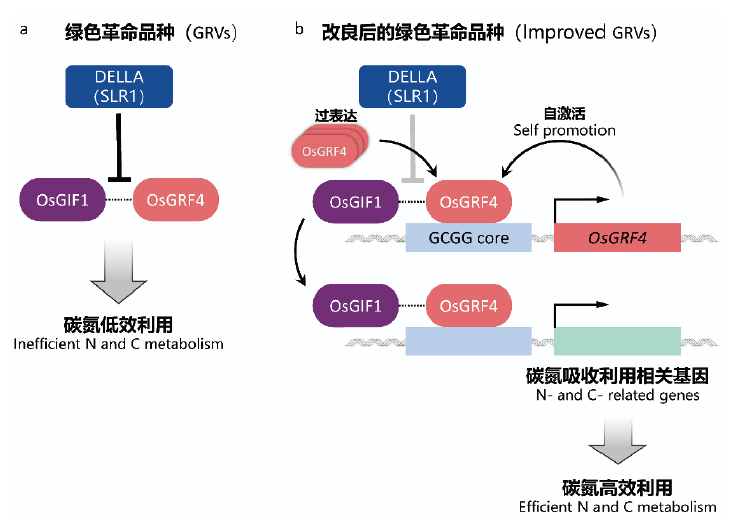
图4 GRF4提高水稻中碳氮代谢效率 a:现有“绿色革命”品种(Green Revolution varieties,GRVs)中,DELLA类蛋白SLR1(SLENDER RICE1)会抑制OsGIF1和OsGRF4的相互作用,影响碳氮代谢;b:改良后的GRVs中,OsGRF4表达量的增加克服了SLR1对OsGIF1和OsGRF4相互作用的抑制,而且OsGRF4通过自激活效应有效的提高了碳氮代谢效率,促进了产量的提升。此图修改自Wang等[60]
Fig. 4 GRF4 increases the efficiency of nitrogen and carbon metabolism in rice a:In Green Revolution varieties(GRVs),DELLA-like protein SLR1(SLENDER RICE1)inhibits the interaction between OsGIF1 and OsGRF4,which affects the metabolism of nitrogen(N)and carbon(C). b:In improved GRVs,increased expressions of OsGRF4 overcomes the inhibition of SLR1 to OsGIF1-OsGRF4 interactions. In addition,OsGRF4 efficiently promotes nitrogen and carbon use efficiency by self-promotion and thus resulting in higher yields. This figure was modified from Wang et al[60]
图5 NGR5基因影响水稻分蘖数量的调控机理 水稻转录因子NGR5促进氮依赖的PRC2募集,进而阻断抑制分蘖基因的表达,促进分蘖。NGR5与赤霉素受体GID1结合后可被降解,而DELLA的积累可竞争性地与GA-GID1结合,抑制GID1-NGR5的相互作用,从而稳定NGR5。此图修改自Wu等[59]5 赤霉素代谢调控与“绿色革命”品种的广适性
Fig. 5 Mechanism of NGR5 regulating rice tillering The rice transcription factor NGR5 facilitates nitrogen-dependent recruitment of PRC2 to repress expression of tillering genes,thus promoting tillering in response to increasing nitrogen supply. NGR5 can be degraded after binding with the gibberellin receptor GID1,while DELLA accumulation inhibits the GID1-NGR5 interaction via competitively binding with GA-GID1,thus stabilizing. This figure was modified from Wu et al[59]
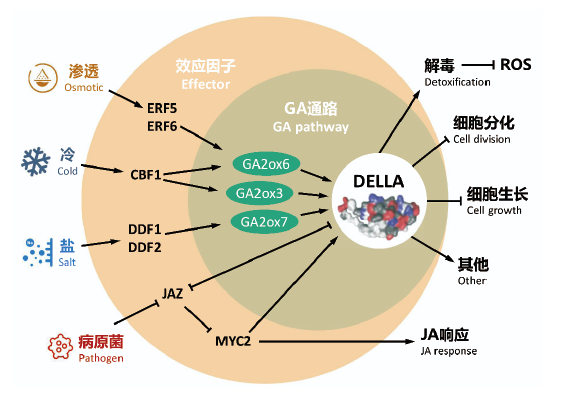
图6 赤霉素参与不同胁迫响应的示意图 不同的非生物胁迫会通过不同的效应因子诱导GA2oxs的表达,导致GAs合成的减少,进而促进DELLA的积累。在病原菌侵染条件下,茉莉酸(jasmonic acid,JA)介导的JAZ蛋白降解会解除其对DELLAs和MYC转录因子的抑制。MYC2同时也会促进DELLA的表达。DELLAs的积累通过抑制细胞伸长和分裂导致细胞停止生长;通过促进解毒酶编码基因表达抑制ROS产生;通过直接抑制JAZ活性促进JA应答。箭头和T条分别表示正调节和负调节。此图修改自Hedden等[1]
Fig.e 6 A simplified model for the role of GAs in stress response Different abiotic stresses induce the expression of GA2oxs via specific effectors,which results in the subsequent decrease of GAs synthesis,thus promotes DELLA accumulation. Under pathogen attack,JA-mediated JAZ protein degradation releases its repression to DELLAs and MYC transcription factors. MYC2 in turn also enhances the expression of DELLA. Accumulation of DELLAs leads to growth cessation via inhibition of cell elongation and cell division. Other protective mechanisms include the expression promotion of genes encoding detoxification enzymes to inhibit ROS generation,and promotion of JA response via direct repression of JAZ activity. Arrows and T-bars indicate positive or negative regulation,respectively. This figure was modified from Hedden et al[1]
| [1] | Hedden P, Thomas SG. Annual plant reviews, volume 49[M]. Chichester, UK:John Wiley & Sons, Ltd, 2016. |
| [2] |
Yamaguchi S. Gibberellin metabolism and its regulation[J]. Annu Rev Plant Biol, 2008, 59(1):225-251.
doi: 10.1146/arplant.2008.59.issue-1 URL |
| [3] |
Hedden P, Thomas SG. Gibberellin biosynjournal and its regulation[J]. Biochem J, 2012, 444(1):11-25.
doi: 10.1042/BJ20120245 pmid: 22533671 |
| [4] |
Hedden P, Sponsel V. A century of gibberellin research[J]. J Plant Growth Regul, 2015, 34(4):740-760.
doi: 10.1007/s00344-015-9546-1 URL |
| [5] |
Hedden P. The current status of research on gibberellin biosynjournal[J]. Plant Cell Physiol, 2020, 61(11):1832-1849.
doi: 10.1093/pcp/pcaa092 pmid: 32652020 |
| [6] |
Hedden P, Phillips AL. Gibberellin metabolism:new insights revealed by the genes[J]. Trends Plant Sci, 2000, 5(12):523-530.
pmid: 11120474 |
| [7] |
Hedden P. The genes of the Green Revolution[J]. Trends Genet, 2003, 19(1):5-9.
doi: 10.1016/s0168-9525(02)00009-4 pmid: 12493241 |
| [8] |
Peng J, Richards DE, Hartley NM, et al. ‘Green revolution’ genes encode mutant gibberellin response modulators[J]. Nature, 1999, 400(6741):256-261.
doi: 10.1038/22307 URL |
| [9] |
Sasaki A, Ashikari M, Ueguchi-Tanaka M, et al. Green revolution:a mutant gibberellin-synjournal gene in rice[J]. Nature, 2002, 416(6882):701-702.
doi: 10.1038/416701a URL |
| [10] | 张忠根. 二十世纪世界农业发展模式的演变[J]. 农业经济, 2001(1):1-3. |
| Zhang ZG. The evolution of the world agricultural development model in the 20th Century[J]. Agric Econ, 2001(1):1-3. | |
| [11] |
Silverstone AL, Sun T. Gibberellins and the green revolution[J]. Trends Plant Sci, 2000, 5(1):1-2.
pmid: 10637654 |
| [12] |
Khush GS. Green revolution:the way forward[J]. Nat Rev Genet, 2001, 2(10):815-822.
pmid: 11584298 |
| [13] |
Salamini F. Hormones and the green revolution[J]. Science, 2003, 302(5642):71-72.
doi: 10.1126/science.1090811 URL |
| [14] | 胡瑞法. 农业科技革命:过去和未来[J]. 农业技术经济, 1998(3):2-11, 50. |
| Hu RF. Agricultural science and technology revolution:past and future[J]. J Agrotech Econ, 1998(3):2-11, 50. | |
| [15] |
Davies WP. An historical perspective from the green revolution to the gene revolution[J]. Nutr Rev, 2003, 61(suppl_6):S124-S134.
doi: 10.1301/nr.2003.jun.S124-S134 URL |
| [16] |
Evenson RE. Assessing the impact of the green revolution, 1960 to 2000[J]. Science, 2003, 300(5620):758-762.
pmid: 12730592 |
| [17] |
Helliwell CA, Sullivan JA, Mould RM, et al. A plastid envelope location of Arabidopsis ent-kaurene oxidase links the plastid and endoplasmic Reticulum steps of the gibberellin biosynjournal pathway[J]. Plant J, 2001, 28(2):201-208.
pmid: 11722763 |
| [18] |
Sun TP, Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynjournal[J]. Plant Cell, 1994, 6(10):1509-1518.
pmid: 7994182 |
| [19] |
Nelson DR, Schuler MA, Paquette SM, et al. Comparative genomics of rice and Arabidopsis. analysis of 727 cytochrome P450 genes and Pseudogenes from a monocot and a dicot[J]. Plant Physiol, 2004, 135(2):756-772.
pmid: 15208422 |
| [20] |
Appleford NEJ, Evans DJ, Lenton JR, et al. Function and transcript analysis of gibberellin-biosynthetic enzymes in wheat[J]. Planta, 2006, 223(3):568-582.
pmid: 16160850 |
| [21] |
Itoh H, Ueguchi-Tanaka M, Sentoku N, et al. Cloning and functional analysis of two gibberellin 3 -hydroxylase genes that are differently expressed during the growth of rice[J]. PNAS, 2001, 98(15):8909-8914.
pmid: 11438692 |
| [22] |
Spray CR, Kobayashi M, Suzuki Y, et al. The dwarf-1(dt)mutant of Zea mays blocks three steps in the gibberellin-biosynthetic pathway[J]. PNAS, 1996, 93(19):10515-10518.
pmid: 11607708 |
| [23] |
Pearce S, Huttly AK, Prosser IM, et al. Heterologous expression and transcript analysis of gibberellin biosynthetic genes of grasses reveals novel functionality in the GA3ox family[J]. BMC Plant Biol, 2015, 15:130.
doi: 10.1186/s12870-015-0520-7 URL |
| [24] |
Plackett ARG, Powers SJ, Fernandez-Garcia N, et al. Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs[J]. Plant Cell, 2012, 24(3):941-960.
doi: 10.1105/tpc.111.095109 URL |
| [25] |
Lee DJ, Zeevaart JA. Molecular cloning of GA 2-oxidase3 from spinach and its ectopic expression in Nicotiana sylvestris[J]. Plant Physiol, 2005, 138(1):243-254.
doi: 10.1104/pp.104.056499 URL |
| [26] |
Thomas SG, Phillips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation[J]. PNAS, 1999, 96(8):4698-4703.
pmid: 10200325 |
| [27] |
Schomburg FM, Bizzell CM, Lee DJ, et al. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants[J]. Plant Cell, 2003, 15(1):151-163.
pmid: 12509528 |
| [28] |
Zhu YY, Nomura T, Xu YH, et al. ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice[J]. Plant Cell, 2006, 18(2):442-456.
doi: 10.1105/tpc.105.038455 URL |
| [29] |
Varbanova M, Yamaguchi S, Yang Y, et al. Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2[J]. Plant Cell, 2007, 19(1):32-45.
pmid: 17220201 |
| [30] |
Sun TP. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants[J]. Curr Biol, 2011, 21(9):R338-R345.
doi: 10.1016/j.cub.2011.02.036 URL |
| [31] |
van de Velde K, Ruelens P, Geuten K, et al. Exploiting DELLA signaling in cereals[J]. Trends Plant Sci, 2017, 22(10):880-893.
doi: 10.1016/j.tplants.2017.07.010 URL |
| [32] |
Ueguchi-Tanaka M, Ashikari M, Nakajima M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin[J]. Nature, 2005, 437(7059):693-698.
doi: 10.1038/nature04028 URL |
| [33] |
Griffiths J, Murase K, Rieu I, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis[J]. Plant Cell, 2006, 18(12):3399-3414.
pmid: 17194763 |
| [34] |
Willige BC, Ghosh S, Nill C, et al. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis[J]. Plant Cell, 2007, 19(4):1209-1220.
pmid: 17416730 |
| [35] |
Murase K, Hirano Y, Sun TP, et al. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1[J]. Nature, 2008, 456(7221):459-463.
doi: 10.1038/nature07519 URL |
| [36] |
Sun TP. Gibberellin-GID1-DELLA:a pivotal regulatory module for plant growth and development[J]. Plant Physiol, 2010, 154(2):567-570.
doi: 10.1104/pp.110.161554 URL |
| [37] |
Harberd NP, Belfield E, Yasumura Y. The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism:how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments[J]. Plant Cell, 2009, 21(5):1328-1339.
doi: 10.1105/tpc.109.066969 URL |
| [38] |
Yu ZP, Duan XB, Luo L, et al. How plant hormones mediate salt stress responses[J]. Trends Plant Sci, 2020, 25(11):1117-1130.
doi: 10.1016/j.tplants.2020.06.008 URL |
| [39] |
Claeys H, De Bodt S, Inzé D. Gibberellins and DELLAs:central nodes in growth regulatory networks[J]. Trends Plant Sci, 2014, 19(4):231-239.
doi: 10.1016/j.tplants.2013.10.001 URL |
| [40] |
Zentella R, Zhang ZL, Park M, et al. Global analysis of della direct targets in early gibberellin signaling in Arabidopsis[J]. Plant Cell, 2007, 19(10):3037-3057.
doi: 10.1105/tpc.107.054999 pmid: 17933900 |
| [41] |
Davière JM, Achard P. A pivotal role of DELLAs in regulating multiple hormone signals[J]. Mol Plant, 2016, 9(1):10-20.
doi: 10.1016/j.molp.2015.09.011 URL |
| [42] |
Law CN, Snape JW, Worland AJ. The genetical relationship between height and yield in wheat[J]. Heredity, 1978, 40(1):133-151.
doi: 10.1038/hdy.1978.13 URL |
| [43] |
Griffiths S, Simmonds J, Leverington M, et al. Meta-QTL analysis of the genetic control of crop height in elite European winter wheat germplasm[J]. Mol Breed, 2012, 29(1):159-171.
doi: 10.1007/s11032-010-9534-x URL |
| [44] |
Peng JH, Sun DF, Nevo E. Domestication evolution, genetics and genomics in wheat[J]. Mol Breed, 2011, 28(3):281-301.
doi: 10.1007/s11032-011-9608-4 URL |
| [45] |
Wilhelm EP, Boulton MI, Barber TES, et al. Genotype analysis of the wheat semidwarfRht-B1bandRht-D1bancestral lineage[J]. Plant Breed, 2013, 132(6):539-545.
doi: 10.1111/pbr.12099 URL |
| [46] |
Guedira M, Brown-Guedira G, van Sanford D, et al. Distribution of rht genes in modern and historic winter wheat cultivars from the eastern and central USA[J]. Crop Sci, 2010, 50(5):1811-1822.
doi: 10.2135/cropsci2009.10.0626 URL |
| [47] |
Radley M. Comparison of endogenous gibberellins and response to applied gibberellin of some dwarf and tall wheat cultivars[J]. Planta, 1970, 92(4):292-300.
doi: 10.1007/BF00385096 pmid: 24500299 |
| [48] |
Gale MD, Marshall GA. Insensitivity to gibberellin in dwarf wheats[J]. Ann Bot, 1973, 37(4):729-735.
doi: 10.1093/oxfordjournals.aob.a084741 URL |
| [49] |
Pearce S, Saville R, Vaughan SP, et al. Molecular characterization of Rht-1 dwarfing genes in hexaploid wheat[J]. Plant Physiol, 2011, 157(4):1820-1831.
doi: 10.1104/pp.111.183657 URL |
| [50] |
Li Y, Xiao J, Wu J, et al. A tandem segmental duplication(TSD)in green revolution gene Rht-D1b region underlies plant height variation[J]. New Phytol, 2012, 196(1):282-291.
doi: 10.1111/nph.2012.196.issue-1 URL |
| [51] |
Wen W, Deng Q, Jia H, et al. Sequence variations of the partially dominant DELLA gene Rht-B1c in wheat and their functional impacts[J]. J Exp Bot, 2013, 64(11):3299-3312.
doi: 10.1093/jxb/ert183 pmid: 23918966 |
| [52] | Rutger JN. Applications of induced and spontaneous mutation in rice breeding and genetics[J]. Adv Agron, 1983, 36:383-413. |
| [53] |
Monna L, Kitazawa N, Yoshino R, et al. Positional cloning of rice semidwarfing gene, sd-1:rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synjournal[J]. DNA Res, 2002, 9(1):11-17.
pmid: 11939564 |
| [54] |
Spielmeyer W, Ellis MH, Chandler PM. Semidwarf(sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene[J]. PNAS, 2002, 99(13):9043-9048.
pmid: 12077303 |
| [55] |
Eshed Y, Lippman ZB. Revolutions in agriculture chart a course for targeted breeding of old and new crops[J]. Science, 2019, 366(6466):eaax0025.
doi: 10.1126/science.aax0025 URL |
| [56] |
Li S, Tian Y, Wu K, et al. Modulating plant growth-metabolism coordination for sustainable agriculture[J]. Nature, 2018, 560(7720):595-600.
doi: 10.1038/s41586-018-0415-5 URL |
| [57] |
Zhang Y, Liu Z, Liu J, et al. GA-DELLA pathway is involved in regulation of nitrogen deficiency-induced anthocyanin accumulation[J]. Plant Cell Rep, 2017, 36(4):557-569.
doi: 10.1007/s00299-017-2102-7 URL |
| [58] |
Liu H, Zhang C, Yang J, et al. Hormone modulation of legume-rhizobial symbiosis[J]. J Integr Plant Biol, 2018, 60(8):632-648.
doi: 10.1111/jipb.v60.8 URL |
| [59] |
Wu K, Wang S, Song W, et al. Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice[J]. Science, 2020, 367(6478):eaaz2046-648.
doi: 10.1126/science.aaz2046 URL |
| [60] | Wang FM, Matsuoka M. Improved nutrient use gives cereal crops a boost[J]. Nature, 2018, 560(7720):563-564. |
| [61] |
Achard P, Cheng H, De Grauwe L, et al. Integration of plant responses to environmentally activated phytohormonal signals[J]. Science, 2006, 311(5757):91-94.
doi: 10.1126/science.1118642 URL |
| [62] |
Magome H, Yamaguchi S, Hanada A, et al. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis[J]. Plant J, 2008, 56(4):613-626.
doi: 10.1111/tpj.2008.56.issue-4 URL |
| [63] |
Achard P, Gong F, Cheminant S, et al. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism[J]. Plant Cell, 2008, 20(8):2117-2129.
doi: 10.1105/tpc.108.058941 URL |
| [64] |
Dubois M, Skirycz A, Claeys H, et al. Ethylene Response Factor6 Acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis[J]. Plant Physiol, 2013, 162(1):319-332.
doi: 10.1104/pp.113.216341 URL |
| [65] |
Kang HG, Kim J, Kim B, et al. Overexpression of FTL1/DDF1, an AP2 transcription factor, enhances tolerance to cold, drought, and heat stresses in Arabidopsis thaliana[J]. Plant Sci, 2011, 180(4):634-641.
doi: 10.1016/j.plantsci.2011.01.002 URL |
| [66] |
Shan C, Mei Z, Duan J, et al. OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress[J]. PLoS One, 2014, 9(1):e87110.
doi: 10.1371/journal.pone.0087110 URL |
| [67] |
Lo SF, Ho TD, Liu YL, et al. Ectopic expression of specific GA2 oxidase mutants promotes yield and stress tolerance in rice[J]. Plant Biotechnol J, 2017, 15(7):850-864.
doi: 10.1111/pbi.12681 URL |
| [68] |
Wang D, Pan Y, Zhao X, et al. Genome-wide temporal-spatial gene expression profiling of drought responsiveness in rice[J]. BMC Genomics, 2011, 12:149.
doi: 10.1186/1471-2164-12-149 pmid: 21406116 |
| [69] |
Kuroha T, Nagai K, Gamuyao R, et al. Ethylene-gibberellin signaling underlies adaptation of rice to periodic flooding[J]. Science, 2018, 361(6398):181-186.
doi: 10.1126/science.aat1577 URL |
| [70] |
Nagai K, Mori Y, Ishikawa S, et al. Antagonistic regulation of the gibberellic acid response during stem growth in rice[J]. Nature, 2020, 584(7819):109-114.
doi: 10.1038/s41586-020-2501-8 URL |
| [71] |
Krugman T, Peleg Z, Quansah L, et al. Alteration in expression of hormone-related genes in wild emmer wheat roots associated with drought adaptation mechanisms[J]. Funct Integr Genom, 2011, 11(4):565-583.
doi: 10.1007/s10142-011-0231-6 URL |
| [72] |
Zhou Y, Underhill SJ. Breadfruit(Artocarpus altilis)gibberellin 2-oxidase genes in stem elongation and abiotic stress response[J]. Plant Physiol Biochem, 2016, 98:81-88.
doi: 10.1016/j.plaphy.2015.11.012 URL |
| [73] |
Zawaski C, Busov VB. Roles of gibberellin catabolism and signaling in growth and physiological response to drought and short-day photoperiods in Populus trees[J]. PLoS One, 2014, 9(1):e86217.
doi: 10.1371/journal.pone.0086217 URL |
| [74] |
Shan XH, Li YD, Jiang Y, et al. Transcriptome profile analysis of maize seedlings in response to high-salinity, drought and cold stresses by deep sequencing[J]. Plant Mol Biol Report, 2013, 31(6):1485-1491.
doi: 10.1007/s11105-013-0622-z URL |
| [75] |
Gaion LA, Monteiro CC, Cruz FJR, et al. Constitutive gibberellin response in grafted tomato modulates root-to-shoot signaling under drought stress[J]. J Plant Physiol, 2018, 221:11-21.
doi: 10.1016/j.jplph.2017.12.003 URL |
| [76] |
Chen L, Phillips AL, Condon AG, et al. GA-responsive dwarfing gene Rht12 affects the developmental and agronomic traits in common bread wheat[J]. PLoS One, 2013, 8(4):e62285.
doi: 10.1371/journal.pone.0062285 URL |
| [77] |
Rebetzke GJ, Richards RA, Fischer VM, et al. Breeding long coleoptile, reduced height wheats[J]. Euphytica, 1999, 106(2):159-168.
doi: 10.1023/A:1003518920119 URL |
| [78] |
Oikawa T, Koshioka M, Kojima K, et al. A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice[J]. Plant Mol Biol, 2004, 55(5):687-700.
doi: 10.1007/s11103-004-1692-y URL |
| [79] |
Itoh H, Ueguchi-Tanaka M, Sakamoto T, et al. Modification of rice plant height by suppressing the height-controlling gene, D18, in rice[J]. Breed Sci, 2002, 52(3):215-218.
doi: 10.1270/jsbbs.52.215 URL |
| [80] |
Hu X, Cui Y, Dong G, et al. Using CRISPR-Cas9 to generate semi-dwarf rice lines in elite landraces[J]. Sci Rep, 2019, 9(1):19096.
doi: 10.1038/s41598-019-55757-9 URL |
| [1] | 张道磊, 甘雨军, 乐亮, 普莉. 玉米产量性状的表观遗传调控机制和育种应用[J]. 生物技术通报, 2023, 39(8): 31-42. |
| [2] | 叶云芳, 田清尹, 施婷婷, 王亮, 岳远征, 杨秀莲, 王良桂. 植物中β-紫罗兰酮生物合成及调控研究进展[J]. 生物技术通报, 2023, 39(8): 91-105. |
| [3] | 王玲, 卓燊, 付学森, 刘紫璇, 刘笑蓉, 王志辉, 周日宝, 刘湘丹. 莲生物碱生物合成途径及相关基因研究进展[J]. 生物技术通报, 2023, 39(7): 56-66. |
| [4] | 成婷, 苑帅, 张晓元, 林良才, 李欣, 张翠英. 酿酒酵母异丁醇合成途径调控的研究进展[J]. 生物技术通报, 2023, 39(7): 80-90. |
| [5] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [6] | 周定定, 李辉虎, 汤兴涌, 余发新, 孔丹宇, 刘毅. 甘草酸和甘草苷生物合成与调控的研究进展[J]. 生物技术通报, 2023, 39(5): 44-53. |
| [7] | 郁慧丽, 李爱涛. 细胞色素P450酶在香精香料绿色生物合成中的应用[J]. 生物技术通报, 2023, 39(4): 24-37. |
| [8] | 金云倩, 王彬, 郭书磊, 赵霖熙, 韩赞平. 赤霉素调控玉米种子活力的研究进展[J]. 生物技术通报, 2023, 39(1): 84-94. |
| [9] | 江美彦, 周杨, 刘仁浪, 姚菲, 杨云舒, 侯凯, 冯冬菊, 吴卫. 白芷根际促生菌的筛选及其促生效果研究[J]. 生物技术通报, 2022, 38(8): 167-178. |
| [10] | 段玥彤, 王鹏年, 张春宝, 林春晶. 植物黄烷酮-3-羟化酶基因研究进展[J]. 生物技术通报, 2022, 38(6): 27-33. |
| [11] | 田清尹, 岳远征, 申慧敏, 潘多, 杨秀莲, 王良桂. 植物观赏器官中类胡萝卜素代谢调控的研究进展[J]. 生物技术通报, 2022, 38(12): 35-46. |
| [12] | 姚宇, 顾佳珺, 孙超, 申国安, 郭宝林. 植物类黄酮UDP-糖基转移酶研究进展[J]. 生物技术通报, 2022, 38(12): 47-57. |
| [13] | 赵玉雪, 王芸, 余璐瑶, 刘京晶, 斯金平, 张新凤, 张磊. 植物中C-糖基转移酶的结构与应用[J]. 生物技术通报, 2022, 38(10): 18-28. |
| [14] | 山琦, 贾惠舒, 姚文博, 刘伟灿, 李海燕. 植物miR396-GRF模块的生物学功能及其潜在应用价值[J]. 生物技术通报, 2022, 38(10): 34-44. |
| [15] | 徐圆圆, 赵国春, 郝颖颖, 翁学煌, 陈仲, 贾黎明. 无患子RT-qPCR内参基因的筛选与验证[J]. 生物技术通报, 2022, 38(10): 80-89. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
