








生物技术通报 ›› 2022, Vol. 38 ›› Issue (5): 175-182.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1039
李豫1( ), 陈刚1(
), 陈刚1( ), 马骞1(
), 马骞1( ), 邝杰华1, 陈有铭2
), 邝杰华1, 陈有铭2
收稿日期:2021-08-16
出版日期:2022-05-26
发布日期:2022-06-10
作者简介:李豫,女,硕士研究生,研究方向:鱼类生物学与遗传育种;E-mail: 基金资助:
LI Yu1( ), CHEN Gang1(
), CHEN Gang1( ), MA Qian1(
), MA Qian1( ), KUANG Jie-hua1, CHEN You-ming2
), KUANG Jie-hua1, CHEN You-ming2
Received:2021-08-16
Published:2022-05-26
Online:2022-06-10
摘要:
旨在探讨军曹鱼nanos1基因(Rcnanos1)的序列特征,及其在胚胎和性腺发育过程中的功能,利用cDNA末端快速扩增技术(RACE)克隆获得Rcnanos1基因的cDNA序列,全长1 290 bp,其中5'非编码区139 bp,3'非编码区470 bp,开放阅读框(ORF)681 bp,共编码226个氨基酸。序列多重比对结果显示,RcNanos1氨基酸序列包含两个连续的锌指功能结构域,与尖吻鲈(Lates calcarifer)Nanos1的一致性最高(99.6%)。系统进化分析表明军曹鱼Nanos1与尖吻鲈(Lates calcarifer)和斜带石斑鱼(Epinephelus coioides)的同源蛋白亲缘关系最近。实时荧光定量PCR(qRT-PCR)结果显示,Rcnanos1在150 dph(days post hatching)军曹鱼的13种组织中均有表达,但在性腺中的表达量显著高于其它组织。胚胎发育过程中,Rcnanos1在早期卵裂阶段的表达水平较低,16细胞期时表达量开始显著升高,并在1 dph仔鱼中的表达量上升至最大值。Rcnanos1在精巢和卵巢首周年发育过程中的表达模式相似,在精巢中(Ⅱ-Ⅴ期),Rcnanos1的表达水平呈逐渐升高趋势,360 dph(Ⅴ期)时的表达量最高;在卵巢中(Ⅰ-Ⅲ期),Rcnanos1的表达量同样在360 dph(Ⅲ期)时达到峰值。由此推测,Rcnanos1基因可能在胚胎和性腺发育过程发挥一定的调控作用,可为揭示军曹鱼生殖细胞发生发育的分子机理提供理论基础。
李豫, 陈刚, 马骞, 邝杰华, 陈有铭. 军曹鱼nanos1的克隆及其在胚胎和性腺发育过程中的表达分析[J]. 生物技术通报, 2022, 38(5): 175-182.
LI Yu, CHEN Gang, MA Qian, KUANG Jie-hua, CHEN You-ming. Cloning of nanos1 Gene of Rachycentron canadum and Its Expression Analysis in the Embryonic and Gonadal Development[J]. Biotechnology Bulletin, 2022, 38(5): 175-182.
| 引物名称 Primer | 序列Sequence(5'-3') | 用途 Usage |
|---|---|---|
| Rcnanos1-F | ATGGATTTTCTCAACCACAACTATT | CDS序列克隆 CDS sequence cloning |
| Rcnanos1-R | TTAGAATATTTTCATCCTCTTACCACC | |
| Rcnanos1-3' F1 | GCCCAAAATCTGCGTCTTCTGC | 3' 末端序列克隆 3' RACE |
| Rcnanos1-3' F2 | TCAAAAGACCAGCCATCCCAGC | |
| Rcnanos1-5' R1 | GCTCCTGGAAGCGGGTTTGC | 5' 末端序列克隆 5' RACE |
| Rcnanos1-5' R2 | CGATTACGCACGGACACGCC | |
| Rcnanos1-F1 | AGAACCCCAACTCCATCACC | Rcnanos1的qRT-PCR检测 Expression ofRcnanos1 |
| Rcnanos1-R1 | TCTGGAAGGGGCTCAGTATC | |
| β-actin-F | AGGGAAATTGTGCGTGAC | 内参基因 Internal control gene |
| β-actin-R | AGGCAGCTCGTAGCTCTT |
表1 本实验所用引物序列
Table 1 Primer sequences used in this study
| 引物名称 Primer | 序列Sequence(5'-3') | 用途 Usage |
|---|---|---|
| Rcnanos1-F | ATGGATTTTCTCAACCACAACTATT | CDS序列克隆 CDS sequence cloning |
| Rcnanos1-R | TTAGAATATTTTCATCCTCTTACCACC | |
| Rcnanos1-3' F1 | GCCCAAAATCTGCGTCTTCTGC | 3' 末端序列克隆 3' RACE |
| Rcnanos1-3' F2 | TCAAAAGACCAGCCATCCCAGC | |
| Rcnanos1-5' R1 | GCTCCTGGAAGCGGGTTTGC | 5' 末端序列克隆 5' RACE |
| Rcnanos1-5' R2 | CGATTACGCACGGACACGCC | |
| Rcnanos1-F1 | AGAACCCCAACTCCATCACC | Rcnanos1的qRT-PCR检测 Expression ofRcnanos1 |
| Rcnanos1-R1 | TCTGGAAGGGGCTCAGTATC | |
| β-actin-F | AGGGAAATTGTGCGTGAC | 内参基因 Internal control gene |
| β-actin-R | AGGCAGCTCGTAGCTCTT |

图1 Rcnanos1 cDNA全长序列和氨基酸序列分析 粗体表示起始密码子和终止密码子;粗体斜体表示加尾信号;灰色阴影表示锌指结构
Fig.1 Complete sequence of Rcnanos1 cDNA and analysis of deduced amino acid sequence The initiation codon ATG and stop codon TAA are in bold. The polyadenylation signal ATTAAA are in bold and italic. The zinc finger motif are highlighted in grey shadow

图2 军曹鱼nanos1氨基酸序列多重序列比对分析 方框表示保守的锌指结构域
Fig.2 Multiple alignment of R. canadum nanos1 deduced amino acid sequence The frame regions indicate the zinc finger motif
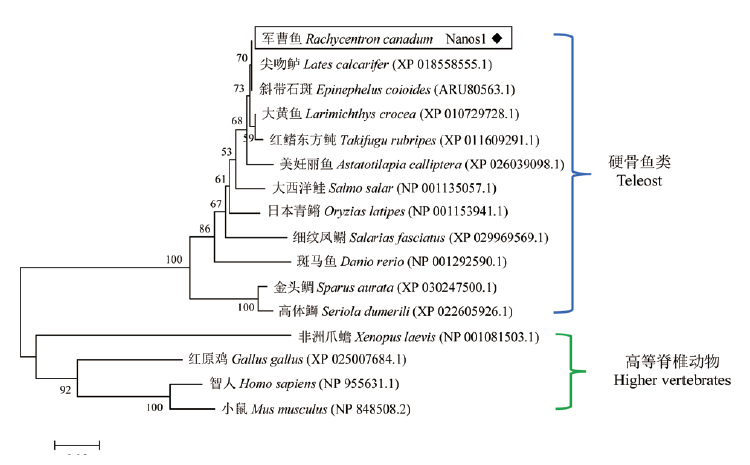
图3 基于nanos1氨基酸序列构建的系统进化树(NJ树) 节点上的数字为各分支的置信度,标尺0.05为进化距离
Fig.3 Phylogenetic tree of nanos1 amino acid sequences based on Neighbor-Joining(NJ)method Numbers indicates the confidence level of each branch,the scale bar 0.05 in terms of genetic distance is indicated below the tree

图4 qRT-PCR分析Rcnanos1在军曹鱼不同组织中的表达 LI:肝脏;SP:脾;KI:体肾;BR:脑;HE:心脏;GI:鳃;TE:精巢;OV:卵巢;ST:胃;IN:肠;MU:肌肉;SK:皮肤;EY:眼睛;上标不同字母表示差异显著(P<0.05)
Fig.4 Expressions of Rcnanos1 in the tissues of R. canadum by qRT-PCR LI:Liver. SP:Spleen. KI:Kidney. BR:Brain. HE:Heart. GI:Gill. TE:Testis. OV:Ovary. ST:Stomach. IN:Intestines. MU:Muscle. SK:Skin. EY:Eye. Different superscripts indicate significant difference(P<0.05)
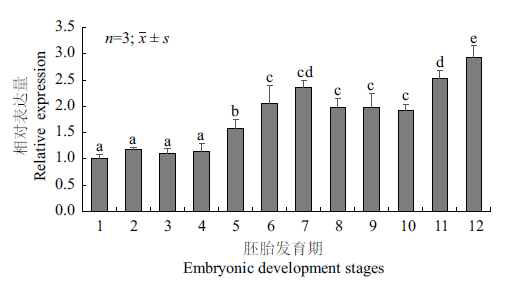
图5 Rcnanos1在胚胎发育过程中的表达分析 1:1细胞期;2:2细胞期;3:4细胞期;4:8细胞期;5:16细胞期;6:多细胞期;7:囊胚期;8:原肠胚期;9:神经胚期;10:器官形成期;11:孵化期;12:孵化后1 d;上标不同字母表示差异显著(P<0.05)
Fig.5 Expressions of Rcnanos1 during the embryonic deve-lopment stages 1:1-cell stage;2:2-cell stage;3:4-cell stage;4:8-cell stage;5:16-cell stage;6:poly-cell stage;7:blastocyst stage;8:gastrulation stage;9:neuroblast stage;10:organ formation stage;11:hatching stage;12:1 day after hatching. Different superscripts indicate significant difference(P<0.05)
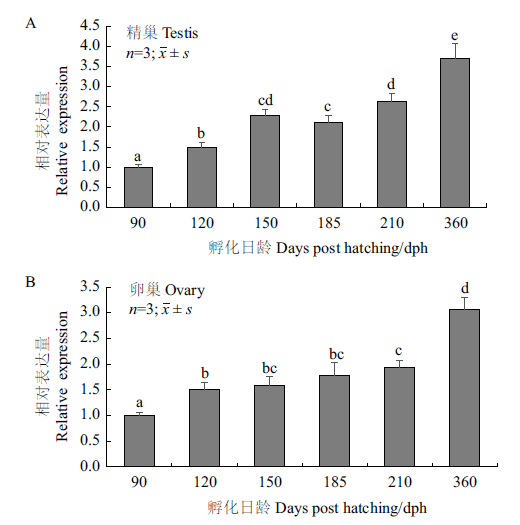
图6 军曹鱼性腺周年发育过程中Rcnanos1 mRNA的表达水平 A:Rcnanos1 mRNA在精巢中的表达水平;B:Rcnanos1 mRNA在卵巢中的表达水平;上标不同字母表示差异显著(P<0.05)
Fig.6 Rcnanos1 mRNA expression in annual gonadal deve-lopment of R. Canadum A:Rcnanos1 mRNA expression in testis;B:Rcnanos1 mRNA expression in ovary. Different superscripts indicate significant difference(P<0.05)
| [1] | 程琳, 黄天晴, 刘晨斌, 等. 鱼类原始生殖细胞标记基因研究进展[J]. 水产学杂志, 2020, 33(6):80-88. |
| Cheng L, Huang TQ, Liu CB, et al. Research perspectives:marker genes of primordial germ cells in fishes[J]. Chin J Fish, 2020, 33(6):80-88. | |
| [2] |
Xu H, Gui J, Hong Y. Differential expression of Vasa RNA and protein during spermatogenesis and oogenesis in the gibel carp(Carassius auratus gibelio), a bisexually and gynogenetically reproducing vertebrate[J]. Dev Dyn, 2005, 233(3):872-882.
doi: 10.1002/dvdy.20410 URL |
| [3] |
Liu L, Hong N, Xu H, et al. Medaka dead end encodes a cytoplasmic protein and identifies embryonic and adult germ cells[J]. Gene Expr Patterns, 2009, 9(7):541-548.
doi: 10.1016/j.gep.2009.06.008 URL |
| [4] |
Sun ZH, Wang Y, Lu WJ, et al. Divergent expression patterns and function implications of four Nanos genes in a hermaphroditic fish, Epinephelus coioides[J]. Int J Mol Sci, 2017, 18(4):685.
doi: 10.3390/ijms18040685 URL |
| [5] |
Bhat N, Hong Y. Cloning and expression of boule and dazl in the Nile tilapia(Oreochromis niloticus)[J]. Gene, 2014, 540(2):140-145.
doi: 10.1016/j.gene.2014.02.057 URL |
| [6] |
Shen R, Xie T. NANOS:a germline stem cell’s Guardian Angel[J]. J Mol Cell Biol, 2010, 2(2):76-77.
doi: 10.1093/jmcb/mjp043 pmid: 20008335 |
| [7] |
Ye H, Chen X, Wei Q, et al. Molecular and expression characterization of a nanos1 homologue in Chinese sturgeon, Acipenser sinensis[J]. Gene, 2012, 511(2):285-292.
doi: 10.1016/j.gene.2012.09.005 URL |
| [8] |
Kadyrova LY, Habara Y, Lee TH, et al. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline[J]. Development, 2007, 134(8):1519-1527.
doi: 10.1242/dev.002212 URL |
| [9] |
Wang Z, Lin H. Nanos maintains germline stem cell self-renewal by preventing differentiation[J]. Science, 2004, 303(5666):2016-2019.
doi: 10.1126/science.1093983 URL |
| [10] | Tan XG, Sui YL, Li MJ, et al. Characterization of nanos1 homolog in the olive flounder, Paralichthys olivaceus(temminck and Schlegel, 1846)[J]. Turk J Fish Aquat Sci, 2020, 20(6):421-429. |
| [11] |
Haraguchi S, Tsuda M, Kitajima S, et al. nanos1:a mouse Nanos gene expressed in the central nervous system is dispensable for normal development[J]. Mech Dev, 2003, 120(6):721-731.
doi: 10.1016/S0925-4773(03)00043-1 URL |
| [12] | 肖懿哲, 朱友芳, 等. 施氏鲟asnanos1基因序列分析及其在雌雄不同组织中表达[J]. 海洋科学, 2017, 41(4):17-23. |
| Xiao YZ, Zhu YF, et al. Sequence and differential expression analyses of the Asnanos1 gene in different tissues of male and female Acipenser schrenckii[J]. Mar Sci, 2017, 41(4):17-23. | |
| [13] |
Köprunner M, Thisse C, Thisse B, et al. A zebrafish Nanos-related gene is essential for the development of primordial germ cells[J]. Genes Dev, 2001, 15(21):2877-2885.
doi: 10.1101/gad.212401 URL |
| [14] |
Zhu W, Wang T, Zhao C, et al. Evolutionary conservation and divergence of Vasa, Dazl and Nanos1 during embryogenesis and gametogenesis in dark sleeper(Odontobutis potamophila)[J]. Gene, 2018, 672:21-33.
doi: 10.1016/j.gene.2018.06.016 URL |
| [15] |
Han K, Chen S, Cai M, et al. Nanos3 not nanos1 and nanos2 is a germ cell marker gene in large yellow croaker during embryogenesis[J]. Comp Biochem Physiol B Biochem Mol Biol, 2018, 218:13-22.
doi: 10.1016/j.cbpb.2018.01.002 URL |
| [16] | 苏敏, 陈元仲, 吕博彦, 等. nanos同源基因在黑脊倒刺鲃生殖细胞中的表达研究[J]. 生物技术, 2012, 22(4):28-32. |
| Su M, et al. Expression of Nanos homolog gene in germ cells of Spinibarbus caldwelli[J]. Biotechnology, 2012, 22(4):28-32. | |
| [17] | Sajeevan MK, Kurup BM. Fecundity and spawning frequency of Cobia, Rachycentron canadum(Linnaeus, 1766)from the North West coast of India[J]. Indian journal of marine sciences, 2016, 45(8):933-936. |
| [18] |
Darden TL, Robinson JD, Strand AE, et al. Forecasting the genetic impacts of net pen failures on gulf of Mexico cobia populations using individual-based model simulations[J]. J World Aquacult Soc, 2017, 48(1):20-34.
doi: 10.1111/jwas.12333 URL |
| [19] | Nguyen MV, Thi Phan LM. Influences of bleeding conditions on the quality and lipid degradation of cobia(Rachycentron canadum)fillets during frozen storage[J]. Turk J Fish and Aquat Sci, 2018, 18(2):289-300. |
| [20] |
Kuo CH, Liao HZ, et al. Highly efficient extraction of EPA/DHA-enriched oil from cobia liver using homogenization plus sonication[J]. Eur J Lipid Sci Technol, 2017, 119(10):1600466.
doi: 10.1002/ejlt.201600466 URL |
| [21] | 刘筠. 中国养殖鱼类繁殖生理学[M]. 北京: 农业出版社, 1993:20-42. |
| Liu Y. Propagation physiology of main cultivated fish in China[M]. Beijing: Agriculture Press, 1993:20-42. | |
| [22] |
Gribouval L, Sourdaine P, et al. The nanos1 gene was duplicated in early Vertebrates and the two paralogs show different gonadal expression profiles in a shark[J]. Sci Rep, 2018, 8(1):6942.
doi: 10.1038/s41598-018-24643-1 pmid: 29720681 |
| [23] |
De Keuckelaere E, et al. Nanos genes and their role in development and beyond[J]. Cell Mol Life Sci, 2018, 75(11):1929-1946.
doi: 10.1007/s00018-018-2766-3 pmid: 29397397 |
| [24] | Lai F. An investigation of Nanos1 function during primordial germ cell development in Xenopus laevis[D]. University of Miami, 2012. |
| [25] |
Hashimoto H, Hara K, Hishiki A, et al. Crystal structure of zinc-finger domain of Nanos and its functional implications[J]. EMBO Rep, 2010, 11(11):848-853.
doi: 10.1038/embor.2010.155 pmid: 20948543 |
| [26] |
Ye B, Petritsch C, Clark IE, et al. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons[J]. Curr Biol, 2004, 14(4):314-321.
doi: 10.1016/j.cub.2004.01.052 URL |
| [27] | 徐钢春, 鲍明明, 等. 鱼类性腺发育及产卵类型研究进展[J]. 长江大学学报:自然科学版, 2017, 14(6):43-48. |
| Xu GC, Bao MM, et al. Research progress on gonadal development and spawning types in fish[J]. J Yangtze Univ:Nat Sci Ed, 2017, 14(6):43-48. | |
| [28] |
Viotti M, Nowotschin S, Hadjantonakis AK. SOX 17 links gut endoderm morphogenesis and germ layer segregation[J]. Nat Cell Biol, 2014, 16(12):1146-1156.
doi: 10.1038/ncb3070 URL |
| [29] | 宋卉, 王树迎. 鱼类原始生殖细胞的研究进展[J]. 动物医学进展, 2004, 25(5):22-23. |
| Song H, Wang SY. Progress in fish primordial germ cells[J]. Prog Vet Med, 2004, 25(5):22-23. |
| [1] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [2] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [3] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [4] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [5] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [6] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [7] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [8] | 王涛, 漆思雨, 韦朝领, 王艺清, 戴浩民, 周喆, 曹士先, 曾雯, 孙威江. CsPPR和CsCPN60-like在茶树白化叶片中的表达分析及互作蛋白验证[J]. 生物技术通报, 2023, 39(3): 218-231. |
| [9] | 庞强强, 孙晓东, 周曼, 蔡兴来, 张文, 王亚强. 菜心BrHsfA3基因克隆及其对高温胁迫的响应[J]. 生物技术通报, 2023, 39(2): 107-115. |
| [10] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| [11] | 葛雯冬, 王腾辉, 马天意, 范震宇, 王玉书. 结球甘蓝PRX基因家族全基因组鉴定与逆境条件下的表达分析[J]. 生物技术通报, 2023, 39(11): 252-260. |
| [12] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [13] | 陈楚怡, 杨小梅, 陈胜艳, 陈斌, 岳莉然. ABA和干旱胁迫下菊花脑ZF-HD基因家族的表达分析[J]. 生物技术通报, 2023, 39(11): 270-282. |
| [14] | 尤垂淮, 谢津津, 张婷, 崔天真, 孙欣路, 臧守建, 武奕凝, 孙梦瑶, 阙友雄, 苏亚春. 钩吻脂氧合酶基因 GeLOX1 的鉴定及低温胁迫表达分析[J]. 生物技术通报, 2023, 39(11): 318-327. |
| [15] | 刘媛媛, 魏传正, 谢永波, 仝宗军, 韩星, 甘炳成, 谢宝贵, 严俊杰. 金针菇II类过氧化物酶基因在子实体发育与胁迫应答过程的表达特征[J]. 生物技术通报, 2023, 39(11): 340-349. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||