








生物技术通报 ›› 2024, Vol. 40 ›› Issue (4): 67-76.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1152
张以恒1,2,3( ), 刘家正4,5,6, 王雪晨2,3, 孙政哲7, 薛雅郡2,3, 汪沛1, 韩华4,5,6, 郑宏伟8(
), 刘家正4,5,6, 王雪晨2,3, 孙政哲7, 薛雅郡2,3, 汪沛1, 韩华4,5,6, 郑宏伟8( ), 李晓娟2,3(
), 李晓娟2,3( )
)
收稿日期:2023-12-08
出版日期:2024-04-26
发布日期:2024-04-30
通讯作者:
郑宏伟,男,博士,研究员,研究方向:生物物理和人工智能;E-mail: hzheng@ms.xjb.ac.cn;作者简介:张以恒,女,硕士研究生,研究方向:生物信息检测与处理、植物细胞生物学;E-mail: zhangyh20@bjfu.edu.cn
基金资助:
ZHANG Yi-heng1,2,3( ), LIU Jia-zheng4,5,6, WANG Xue-chen2,3, SUN Zheng-zhe7, XUE Ya-jun2,3, WANG Pei1, HAN Hua4,5,6, ZHENG Hong-wei8(
), LIU Jia-zheng4,5,6, WANG Xue-chen2,3, SUN Zheng-zhe7, XUE Ya-jun2,3, WANG Pei1, HAN Hua4,5,6, ZHENG Hong-wei8( ), LI Xiao-juan2,3(
), LI Xiao-juan2,3( )
)
Received:2023-12-08
Published:2024-04-26
Online:2024-04-30
摘要:
【目的】为了解决在植物细胞内质网的研究中,成像速度与成像分辨率难以同时满足准确识别精细结构和动态变化的瓶颈问题。【方法】使用结构光照明显微成像技术,对拟南芥活体材料中的内质网进行超分辨实时成像,并优化了自监督去噪框架(Blind2Unblind),以进一步提升快速显微成像的信噪比。【结果】建立了对时间序列成像中内质网结构进行定量分析的方法,并通过对环境胁迫下内质网结构动态变化的追踪进一步验证了方法的有效性。此外,各类参数的相关性分析显示管状内质网的面积和长度与生长端和三叉点的数量显著正相关,而内质网池和整体流的面积与管的面积和长度显著负相关。【结论】优化的自监督去噪框架提升了植物活细胞中结构光照明显微图像的信噪比,实现了管状内质网、内质网池、整体流、生长端和节点等复杂结构和动态的量化,各结构间存在复杂相关性。
张以恒, 刘家正, 王雪晨, 孙政哲, 薛雅郡, 汪沛, 韩华, 郑宏伟, 李晓娟. 基于超分辨成像增强对拟南芥内质网动态变化的研究[J]. 生物技术通报, 2024, 40(4): 67-76.
ZHANG Yi-heng, LIU Jia-zheng, WANG Xue-chen, SUN Zheng-zhe, XUE Ya-jun, WANG Pei, HAN Hua, ZHENG Hong-wei, LI Xiao-juan. Dynamic Changes of Arabidopsis Endoplasmic Reticulum Based on Enhanced Super-resolution Images[J]. Biotechnology Bulletin, 2024, 40(4): 67-76.

图1 基于植物根超分辨成像的去噪增强、自动识别及量化分析流程图 A:以拟南芥幼苗为研究对象,在活体材料中对根细胞中内质网的结构和动态进行研究,方框所示区域为伸长区(Bar=2 μm);B:激光共聚焦显微镜采集的内质网图像;C:使用SIM对内质网进行成像;D:SIM对内质网的成像结果;E:利用增强去噪框架处理后的成像结果;F:利用Swint-ResU-Net对采集图像进行内质网结构特征分割(绿色为整体流;红色为内质网体,深蓝色为管,浅蓝色为池);G:对采集量化的9种特征参数(整体流面积占内质网总面积比例和管面积占内质网总面积比例作为举例)做统计;H:对内质网结构参数进行量化并分析各参数间相关性。
Fig. 1 Workflow for de-noising, enhancement, automatic identification and quantitative analysis based on the super-resolution imaging in the root zone of plant A: The structure and dynamics of ER(endoplasmic reticulum)in the root cells were studied in living material using Arabidopsis seedlings, with the region shown in the box as the elongation zone(Bar = 2 μm). B: ER images acquired by confocal microscopy. C: Imaging of ER using SIM. D: Imaging results of ER using SIM. E: Imaging results processed by utilizing the enhancement denoising framework. F: Segmentation of ER structural features on the acquired images using Swint-ResU-Net(green markers are bulk flow; red markers are fusiform body, darks blue markers are the tubules, and light blue markers are cisternaes). G: Statistics were done on the nine feature parameters quantified by the acquisition(the ratio of the areal bulk flow to the total area of the ER and the ratio of the areal tubule to the total area of the ER as examples). H: Quantitative and correlation analysis of the ER structural parameters

图2 Blind2Unblind去噪框架的概述 A:训练步骤。全局掩码器Ω(·)向噪声图像y添加盲点来创建掩码体块。接着,全局感知掩蔽映射器对去噪体块盲点处采样得到h(fθ(Ωy))。同时,去噪器fθ(·)将y作为输入,输出抑噪结果fθ(y)。重新可见损失以不可见项h(fθ(Ωy))为梯度更新媒介,实现从盲到可见的过渡。此外,正则项被用来稳定训练。B:使用训练的去噪模型进行推理。去噪网络直接从噪声图像y中生成去噪图像,无需额外操作
Fig. 2 Overview of the Blind2Unblind de-noising framework A: Training process. The global masker Ω(·) introduces blind spots into the noisy image y, creating masked patches. Subsequently, the global perc; eptual masking mapper samples h(fθ(Ωy))at the blind spots of the denoised patches. Simultaneously, the denoiser fθ(·) takes y as input and produces the denoised output fθ(y). The re-visible loss employs the imperceptible term h(fθ(Ωy)) as a gradient update medium, facilitating the transition from blind to visible. Additionally, regularization terms are utilized to stabilize the training. B: Inference using the trained denoising model. The denoising network directly generates denoised images from the noisy image y without the need for additional operations.
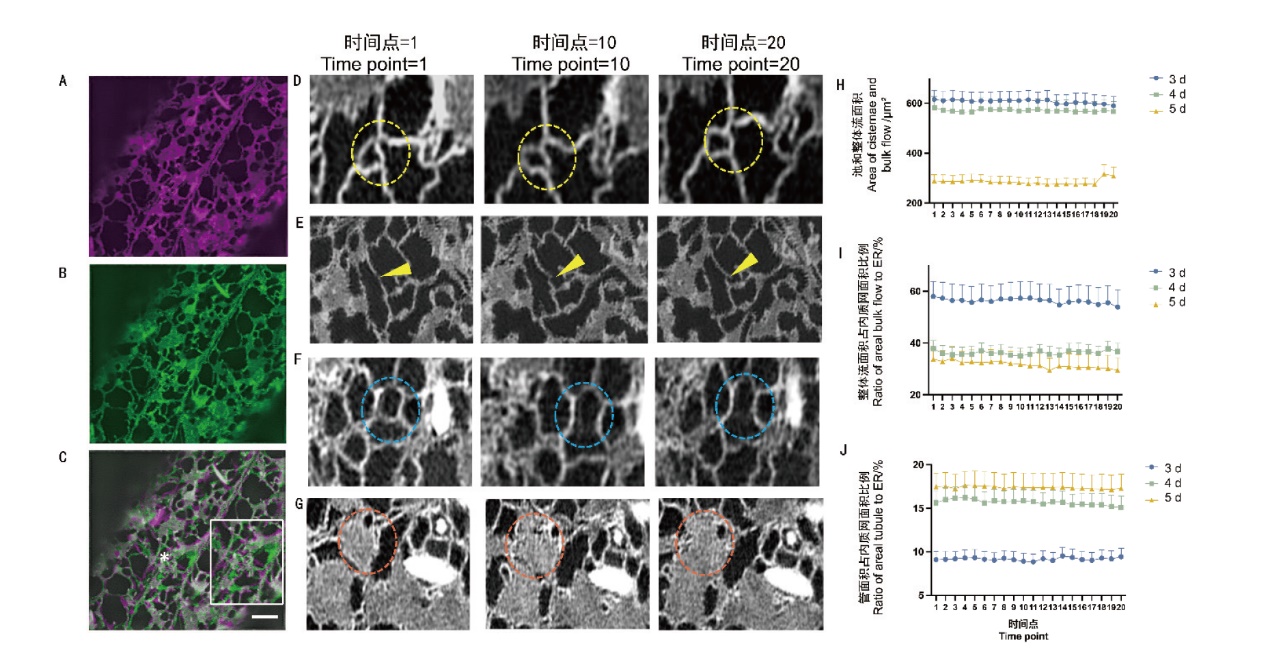
图3 时间序列中内质网的结构变化 A:拍摄时间点为1时的内质网结构成像结果;B:拍摄时间点为20时的内质网结构成像结果;C:A和B的重叠结果,右侧白色方框为星号区域放大后的展示(Bar=2 μm);D:圆圈内的三叉点和多叉点在时间序列内的动态变化;E:箭头指示生长端在时间序列内的动态变化;F:圆圈内的管在时间序列内的动态变化;G:圆圈内的池在时间序列内的动态变化;H:3、4、5 d中内质网池和整体流面积占内质网比例在时间序列下的动态变化;I:3、4、5 d中内质网整体流面积占内质网比例在时间序列下的动态变化;J:3、4、5 d中管面积占内质网面积的比例在时间序列下的动态变化(n=21个细胞)
Fig. 3 Structural changes of ER in the time series A: Imaging results of ER structure when the time point of shooting is 0. B: Imaging results of ER structure when the time point of shooting is 20. C: Overlapping results of A and B. The white box on the right side is the demonstrated zoomed the asterisked area. Bar = 2 μm. D: Dynamic change of the three-way junction and multi-way junction inside the circle inside the time series. E: Arrows indicate the dynamics of the growth tip within the time series. F: Dynamic changes of tubules within circles within the time series. G: Dynamic changes of cisternae within circles within the time series. H: Dynamic changes of ER cisternae and bulk flow area under the time series in day 3, 4, and 5. I: Dynamic changes of ER bulk flow area as a proportion of endoplasmic reticulum under the time series of in day 3, 4, and 5. J: Dynamic changes of areal tubule as a proportion of ER area in day 3, 4, and 5 under the time series(n=21 cells)
| 数据 Data | 3 d | 4 d | 5 d | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 池和整体流面积Area of cisternae and bulk flow/μm2 | 整体流面积占内质网面积比例 Ratio of areal bulk flow to ER/% | 管面积占内质网面积比例Ratio of areal tubule to ER/% | 池和整体流面积Area of cisternae and bulk flow/μm2 | 整体流面积占内质网面积比例Ratio of areal bulk flow to ER/% | 管面积占内质网面积比例Ratio of areal tubule to ER/% | 池和整体流面积Area of cisternae and bulk flow/μm2 | 整体流面积占内质网面积比例Ratio of areal bulk flow to ER/% | 管面积占内质网面积比例Ratio of areal tubule to ER/% | ||||
| 最大值 Maximum | 1 018.63 | 69.76 | 10.98 | 582.88 | 33.67 | 14.27 | 499.43 | 30.70 | 17.13 | |||
| 最小值 Minimum | 236.92 | 66.85 | 9.94 | 32.03 | 32.03 | 13.10 | 486.89 | 27.12 | 16.26 | |||
| 平均数 Mean | 606.85 ±52.46 | 68.71 ±0.78 | 10.38 ±0.06 | 570.93 ±22.58 | 48.71 ±0.72 | 13.65 ±0.12 | 492.25 ±9.92 | 40.36 ±4.26 | 16.79 ±0.05 | |||
表1 时间序列中伸长区细胞各内质网结构参数的统计数据
Table 1 Statistical data of each ER structural parameter of cells in the elongation zone in the time series
| 数据 Data | 3 d | 4 d | 5 d | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 池和整体流面积Area of cisternae and bulk flow/μm2 | 整体流面积占内质网面积比例 Ratio of areal bulk flow to ER/% | 管面积占内质网面积比例Ratio of areal tubule to ER/% | 池和整体流面积Area of cisternae and bulk flow/μm2 | 整体流面积占内质网面积比例Ratio of areal bulk flow to ER/% | 管面积占内质网面积比例Ratio of areal tubule to ER/% | 池和整体流面积Area of cisternae and bulk flow/μm2 | 整体流面积占内质网面积比例Ratio of areal bulk flow to ER/% | 管面积占内质网面积比例Ratio of areal tubule to ER/% | ||||
| 最大值 Maximum | 1 018.63 | 69.76 | 10.98 | 582.88 | 33.67 | 14.27 | 499.43 | 30.70 | 17.13 | |||
| 最小值 Minimum | 236.92 | 66.85 | 9.94 | 32.03 | 32.03 | 13.10 | 486.89 | 27.12 | 16.26 | |||
| 平均数 Mean | 606.85 ±52.46 | 68.71 ±0.78 | 10.38 ±0.06 | 570.93 ±22.58 | 48.71 ±0.72 | 13.65 ±0.12 | 492.25 ±9.92 | 40.36 ±4.26 | 16.79 ±0.05 | |||
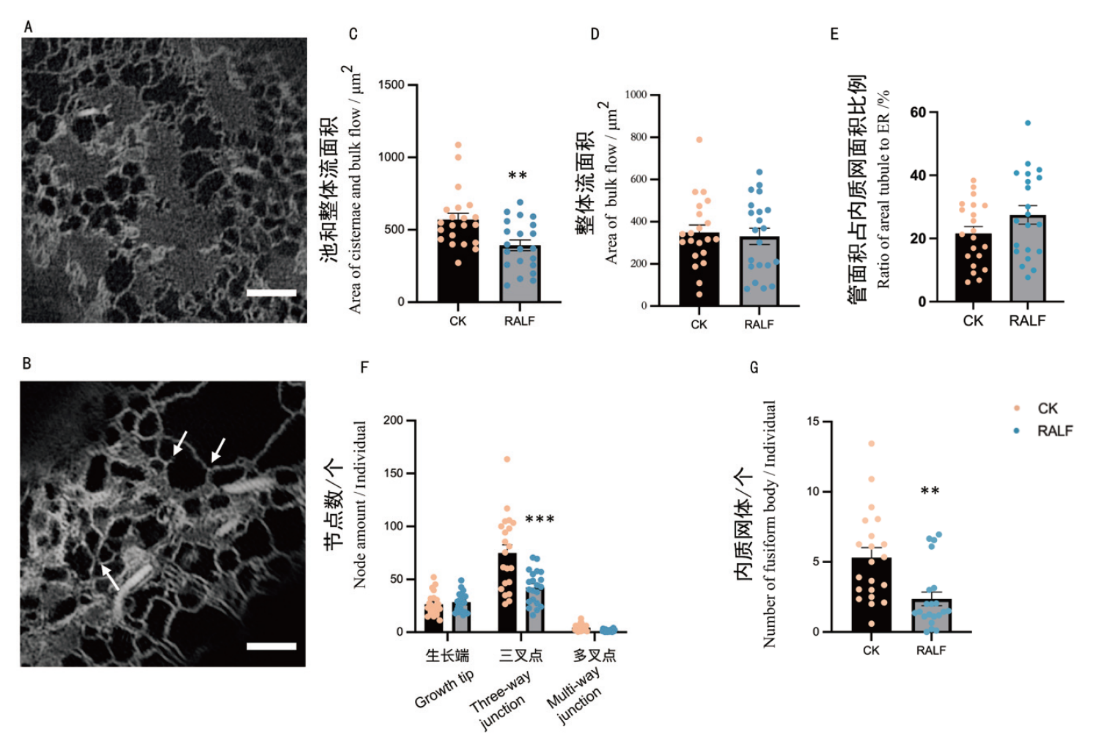
图4 伸长区细胞在第4天以及RALF胁迫条件下的量化分析 A:伸长区细胞正常生长状态下第4天时成像的识别结果;B:在RALF胁迫下,伸长区细胞第4天的成像识别结果(箭头所指示从左往右依次为:多叉点,生长点和三叉点);C:CK与RALF胁迫下,池和整体流面积的量化;D:CK与RALF胁迫下,整体流面积的量化;E. CK与RALF胁迫下,管面积占内质网面积比例的量化;F:CK与RALF胁迫下,节点数的量化;G:CK与RALF胁迫下,内质网体个数的量化。*: P< 0.05,**: P< 0.01, ***: P< 0.001(n=21个细胞)
Fig. 4 Elongation zone cells analyzed at day 4 and under RALF stress conditions A: Identification results of elongation zone cells imaged at day 4 under normal growth conditions. B: Identification results of elongation zone cells imaged at day 4 under RALF stress(From left to right, as indicated by the arrows, the sequence is as follows: multi-way junction, growth tip and three-way junction). C: Quantification of areal cisternae and bulk flow under CK and RALF stress. D: Quantification of areal bulk flow under CK and RALF stress. E: Ratio of areal tubule to areal ER under CK and RALF stress quantification. F: Quantification of the number of nodes under CK and RALF stress. G: Quantification of the number of fusiform body under CK and RALF stress. *: P < 0.05, **:P < 0.01, ***: P< 0.001(n=21 cells)
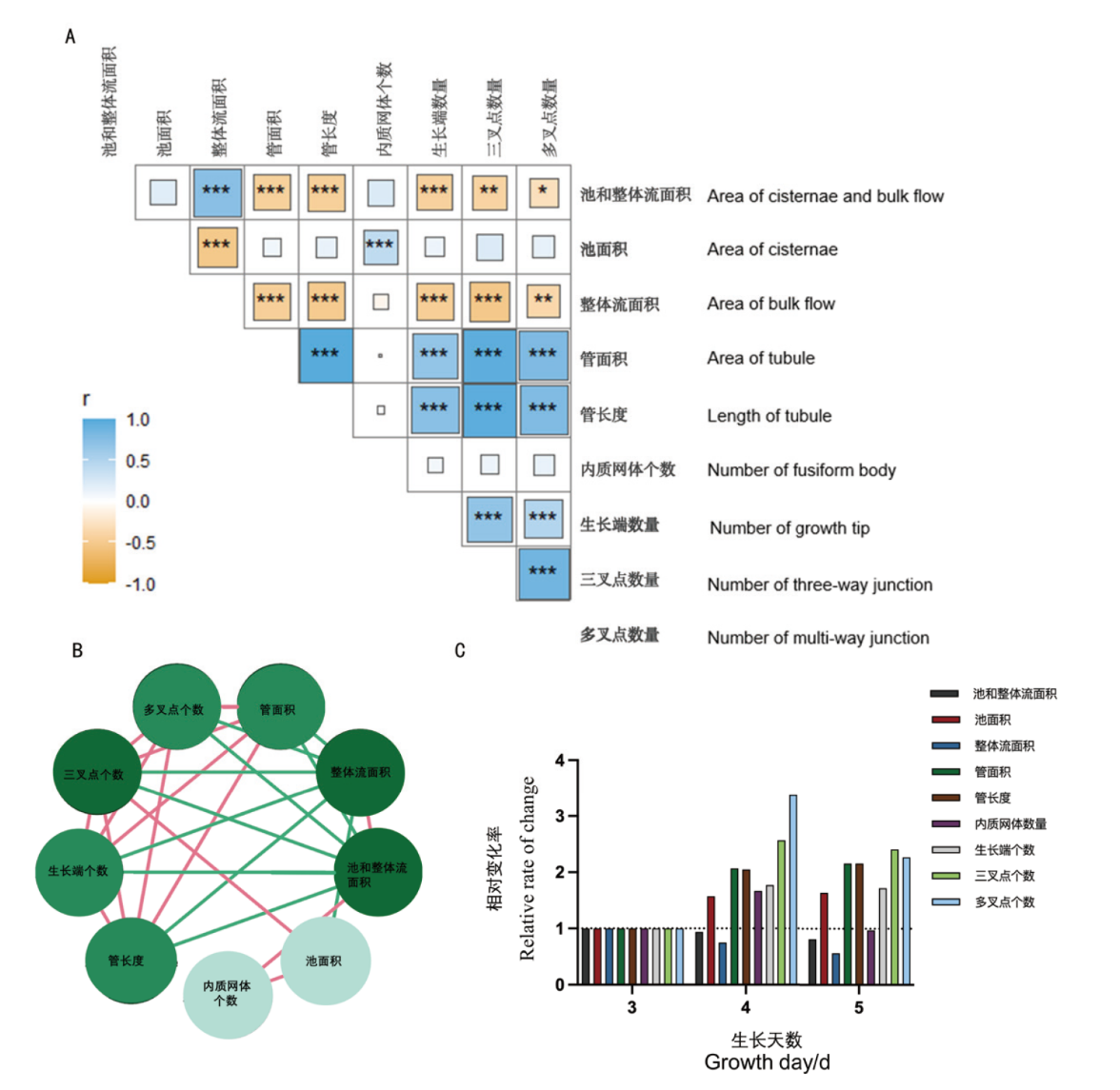
图5 9种特征参数在第3天、4天、5天测量数据的相关性对比图 A:各结构参数在3、4、5 d量化数据的Pearson’s相关性对比图。B:各结构参数之间的相关性图,红线是正相关性,绿线表示负相关性;相关性关系数量与结构参数圆圈颜色呈正比。C:9种参数特征的相对变化率。*: P< 0.05,**: P< 0.01, ***: P< 0.001
Fig. 5 Comparative correlation analysis of nine feature parameters on the day 3, 4, and 5 of measurement A: Pearson's correlation analysis for quantified data of structural parameters on day 3, 4, and 5, respectively. B: The inter-parameter correlations with red lines denoting positive correlations and green lines representing negative correlations; the intensity of correlation is proportional to the circle color. C: The relative rate of change for the nine feature parameters. *: P< 0.05,**: P< 0.01, ***: P< 0.001
| [1] |
Berridge MJ. The endoplasmic reticulum: a multifunctional signaling organelle[J]. Cell Calcium, 2002, 32(5-6): 235-249.
doi: 10.1016/s0143416002001823 pmid: 12543086 |
| [2] |
Staehelin LA. The plant ER: a dynamic organelle composed of a large number of discrete functional domains[J]. Plant J, 1997, 11(6): 1151-1165.
doi: 10.1046/j.1365-313x.1997.11061151.x pmid: 9225461 |
| [3] | English AR, Voeltz GK. Endoplasmic reticulum structure and interconnections with other organelles[J]. Cold Spring Harb Perspect Biol, 2013, 5(4): a013227. |
| [4] |
Anderson DJ, Hetzer MW. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation[J]. J Cell Biol, 2008, 182(5): 911-924.
doi: 10.1083/jcb.200805140 pmid: 18779370 |
| [5] |
Kadowaki H, Nishitoh H. Endoplasmic reticulum quality control by garbage disposal[J]. FEBS J, 2019, 286(2): 232-240.
doi: 10.1111/febs.14589 pmid: 29923316 |
| [6] |
Agellon LB, Michalak M. The endoplasmic reticulum and the cellular reticular network[J]. Adv Exp Med Biol, 2017, 981: 61-76.
doi: 10.1007/978-3-319-55858-5_4 pmid: 29594858 |
| [7] | Lu M, Christensen CN, Weber JM, et al. ERnet: a tool for the semantic segmentation and quantitative analysis of endoplasmic reticulum topology[J]. Nat Methods, 2023, 20(4): 569-579. |
| [8] |
Bravo R, Parra V, Gatica D, et al. Endoplasmic reticulum and the unfolded protein response: dynamics and metabolic integration[J]. Int Rev Cell Mol Biol, 2013, 301: 215-290.
doi: 10.1016/B978-0-12-407704-1.00005-1 pmid: 23317820 |
| [9] |
Baumann O, Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains[J]. Int Rev Cytol, 2001, 205: 149-214.
pmid: 11336391 |
| [10] |
Wozny MR, Di Luca A, Morado DR, et al. In situ architecture of the ER-mitochondria encounter structure[J]. Nature, 2023, 618(7963): 188-192.
doi: 10.1038/s41586-023-06050-3 |
| [11] |
Kornmann B, Currie E, Collins SR, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen[J]. Science, 2009, 325(5939): 477-481.
doi: 10.1126/science.1175088 pmid: 19556461 |
| [12] |
Friedman JR, Lackner LL, West M, et al. ER tubules mark sites of mitochondrial division[J]. Science, 2011, 334(6054): 358-362.
doi: 10.1126/science.1207385 pmid: 21885730 |
| [13] |
Hamasaki M, Furuta N, Matsuda A, et al. Autophagosomes form at ER-mitochondria contact sites[J]. Nature, 2013, 495(7441): 389-393.
doi: 10.1038/nature11910 |
| [14] | Li C, Duckney P, Zhang T. et al. TraB family proteins are components of ER-mitochondrial contact sites and regulate ER-mitochondrial interactions and mitophagy[J]. Nat Commun 13,2022, 13(1):5658. |
| [15] |
Ridge RW, Uozumi Y, Plazinski J, et al. Developmental transitions and dynamics of the cortical ER of Arabidopsis cells seen with green fluorescent protein[J]. Plant Cell Physiol, 1999, 40(12): 1253-1261.
pmid: 10682347 |
| [16] | Stefano G, Brandizzi F. Unique and conserved features of the plant ER-shaping GTPase RHD3[J]. Cell Logist, 2014, 4(1): e28217. |
| [17] |
Voeltz GK, Prinz WA, Shibata Y, et al. A class of membrane proteins shaping the tubular endoplasmic reticulum[J]. Cell, 2006, 124(3): 573-586.
doi: 10.1016/j.cell.2005.11.047 pmid: 16469703 |
| [18] |
De Craene JO, Coleman J, de Martin PE, et al. Rtn1p is involved in structuring the cortical endoplasmic reticulum[J]. Mol Biol Cell, 2006, 17(7): 3009-3020.
doi: 10.1091/mbc.e06-01-0080 URL |
| [19] |
Tolley N, Sparkes IA, Hunter PR, et al. Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport[J]. Traffic, 2008, 9(1): 94-102.
pmid: 17980018 |
| [20] |
West M, Zurek N, Hoenger A, et al. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature[J]. J Cell Biol, 2011, 193(2): 333-346.
doi: 10.1083/jcb.201011039 pmid: 21502358 |
| [21] |
Chang J, Lee S, Blackstone C. Protrudin binds atlastins and endoplasmic reticulum-shaping proteins and regulates network formation[J]. Proc Natl Acad Sci USA, 2013, 110(37): 14954-14959.
doi: 10.1073/pnas.1307391110 pmid: 23969831 |
| [22] | Pain C, Kriechbaumer V, Kittelmann M, et al. Quantitative analysis of plant ER architecture and dynamics[J]. Nat Commun, 2019, 10(1): 984. |
| [23] |
Pearce G, Moura DS, Stratmann J, et al. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development[J]. Proc Natl Acad Sci USA, 2001, 98(22): 12843-12847.
pmid: 11675511 |
| [24] |
Holcman D, Parutto P, Chambers JE, et al. Single particle trajectories reveal active endoplasmic reticulum luminal flow[J]. Nat Cell Biol, 2018, 20(10): 1118-1125.
doi: 10.1038/s41556-018-0192-2 pmid: 30224760 |
| [25] |
Taitt CR, Anderson GP, Ligler FS. Evanescent wave fluorescence biosensors[J]. Biosens Bioelectron, 2005, 20(12):2470-2487.
pmid: 15854820 |
| [26] | Damen D, Hogg DC. Computer vision and pattern recognition(CVPR)[C]. New Orleans:IEEE, 2009 |
| [27] |
Schwarz DS, Blower MD. The endoplasmic reticulum: structure, function and response to cellular signaling[J]. Cell Mol Life Sci, 2016, 73(1): 79-94.
doi: 10.1007/s00018-015-2052-6 pmid: 26433683 |
| [28] | Kim JS, Mochida K, Shinozaki K. ER stress and the unfolded protein response: homeostatic regulation coordinate plant survival and growth[J]. Plants, 2022, 11(23): 3197. |
| [29] | Di Conza G, Ho PC. ER stress responses: an emerging modulator for innate immunity[J]. Cells, 2020, 9(3): 695. |
| [30] |
Zhang LR, Bassham DC, Pittendrigh BR. Editorial: plant ER stress and the UPR signaling pathways[J]. Front Plant Sci, 2022, 13: 968353.
doi: 10.3389/fpls.2022.968353 URL |
| [31] |
Thor F, Gautschi M, Geiger R, et al. Bulk flow revisited: transport of a soluble protein in the secretory pathway[J]. Traffic, 2009, 10(12): 1819-1830.
doi: 10.1111/j.1600-0854.2009.00989.x pmid: 19843282 |
| [1] | 陈春林, 李白雪, 李金玲, 杜清洁, 李猛, 肖怀娟. 甜瓜CmEPF基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(4): 130-138. |
| [2] | 桑森骅. 基于实时动态成像系统对NK细胞毒性的检测方法[J]. 生物技术通报, 2024, 40(4): 77-84. |
| [3] | 付威, 韦素云, 陈赢男. 植物生长发育动态QTL解析研究进展[J]. 生物技术通报, 2024, 40(2): 9-19. |
| [4] | 李宇, 李素贞, 陈茹梅, 卢海强. 植物bHLH转录因子调控铁稳态的研究进展[J]. 生物技术通报, 2023, 39(7): 26-36. |
| [5] | 李帜奇, 袁月, 苗荣庆, 庞秋颖, 张爱琴. 盐胁迫盐芥和拟南芥褪黑素含量及合成相关基因表达模式分析[J]. 生物技术通报, 2023, 39(5): 142-151. |
| [6] | 崔吉洁, 蔡文波, 庄庆辉, 高爱平, 黄建峰, 陈亚辉, 宋志忠. 杧果Fe-S簇装配基因MiISU1的生物学功能[J]. 生物技术通报, 2023, 39(2): 139-146. |
| [7] | 蒋铭轩, 李康, 罗亮, 刘建祥, 芦海平. 植物表达外源蛋白研究进展及展望[J]. 生物技术通报, 2023, 39(11): 110-122. |
| [8] | 鄢梦雨, 韦晓薇, 曹婧, 兰海燕. 异子蓬SabHLH169基因的克隆及抗旱功能分析[J]. 生物技术通报, 2023, 39(11): 328-339. |
| [9] | 阮航, 多浩源, 范文艳, 吕清晗, 姜述君, 朱生伟. AtERF49在拟南芥应答盐碱胁迫中的作用[J]. 生物技术通报, 2023, 39(1): 150-156. |
| [10] | 林蓉, 郑月萍, 徐雪珍, 李丹丹, 郑志富. 拟南芥ACOL8基因在乙烯合成与响应中的功能分析[J]. 生物技术通报, 2023, 39(1): 157-165. |
| [11] | 高聪, 萧楚健, 鲁帅, 王苏蓉, 袁卉华, 曹云英. 氧化石墨烯对拟南芥生长的促进作用[J]. 生物技术通报, 2022, 38(6): 120-128. |
| [12] | 徐红云, 张明意. GRAS转录因子AtSCL4负调控拟南芥应答渗透胁迫[J]. 生物技术通报, 2022, 38(6): 129-135. |
| [13] | 古盼, 齐学影, 李莉, 张曦, 单晓昳. AtRGS1胞吞动态调控G蛋白参与拟南芥生长发育和抗性反应[J]. 生物技术通报, 2022, 38(6): 34-42. |
| [14] | 周娟, 阎晋东, 李新梅, 刘雪晴, 赵强, 赵小英. 拟南芥F-box蛋白FKF1与转录因子FUL互作调控开花研究[J]. 生物技术通报, 2022, 38(3): 1-8. |
| [15] | 杨佳慧, 孙玉萍, 陆雅宁, 刘欢, 卢存福, 陈玉珍. 拟南芥AtTERT对大肠杆菌非生物胁迫抗性的影响[J]. 生物技术通报, 2022, 38(2): 1-9. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||