








生物技术通报 ›› 2025, Vol. 41 ›› Issue (11): 261-271.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0381
• 研究报告 • 上一篇
何文馨1( ), 陈玲1, 黄灵琳1, 徐焱1, 孙祖栋2, 曾维英2, 常小丽1(
), 陈玲1, 黄灵琳1, 徐焱1, 孙祖栋2, 曾维英2, 常小丽1( ), 杨峰1(
), 杨峰1( )
)
收稿日期:2025-04-12
出版日期:2025-11-26
发布日期:2025-12-09
通讯作者:
常小丽,女,副教授,研究方向 :作物高产与抗病协同分子机制;E-mail: xl_chang14042@sicau.edu.cn作者简介:何文馨,女,研究方向 :植物抗病基因挖掘;E-mail: 1392733453@qq.com基金资助:
HE Wen-xin1( ), CHEN Ling1, HUANG Ling-lin1, XU Yan1, SUN Zu-dong2, ZENG Wei-ying2, CHANG Xiao-li1(
), CHEN Ling1, HUANG Ling-lin1, XU Yan1, SUN Zu-dong2, ZENG Wei-ying2, CHANG Xiao-li1( ), YANG Feng1(
), YANG Feng1( )
)
Received:2025-04-12
Published:2025-11-26
Online:2025-12-09
摘要:
目的 探究大豆油菜素内酯信号响应转录因子GmBEE1a和GmBEE3的生物学功能,为大豆抗病与高产基因资源的挖掘提供科学依据。 方法 以南豆12为供试材料,采用生物信息学分析GmBEEs基因结构、编码蛋白的理化性质及其空间构象,克隆GmBEE1a和GmBEE3基因编码序列(CDS)片段,并观察蛋白的亚细胞定位;通过实时定量荧光PCR技术分析GmBEEs基因的组织特异性表达及对不同处理的响应特性。 结果 大豆GmBEE1a和GmBEE3均含有6个外显子和5个内含子,属于典型的bHLH类转录因子,编码酸性不稳定蛋白;系统进化分析揭示GmBEE1a和GmBEE3蛋白与拟南芥、烟草同源蛋白亲缘关系较近;启动子顺式作用元件分析显示,两个基因启动子区域均包含光响应、胁迫应答及激素调控等元件;烟草瞬时表达实验表明,GmBEE1a和GmBEE3蛋白定位在细胞核中。RT-qPCR结果表明,GmBEE1a和GmBEE3在大豆花和叶中的表达水平高于其他组织;根腐病尖孢镰孢菌B3S1和细菌鞭毛蛋白多肽flag22处理均显著诱导2个GmBEEs表达,但根际促生绿针假单胞菌IRHB3处理则抑制其表达;此外,外源水杨酸(SA)、茉莉酸甲酯(MeJA)、油菜素内酯(BL)处理后,GmBEE1a 和 GmBEE3均被显著瞬时诱导表达。 结论 大豆bHLH转录因子GmBEE1a和GmBEE3不仅参与调控大豆生长发育,而且能响应微生物侵染及外源激素调控。
何文馨, 陈玲, 黄灵琳, 徐焱, 孙祖栋, 曾维英, 常小丽, 杨峰. 大豆油菜素内酯信号响应转录因子GmBEE1和GmBEE3的克隆及其表达模式分析[J]. 生物技术通报, 2025, 41(11): 261-271.
HE Wen-xin, CHEN Ling, HUANG Ling-lin, XU Yan, SUN Zu-dong, ZENG Wei-ying, CHANG Xiao-li, YANG Feng. Cloning and Expression Pattern Analysis of Brassinosteriod Signal Response Transcription Factors GmBEE1 and GmBEE3 in Soybean[J]. Biotechnology Bulletin, 2025, 41(11): 261-271.
引物名称 Primer name | 上游引物 Forward primer (5′‒3′) | 下游引物 Reverse primer (5′‒3′) |
|---|---|---|
| GmBEE1a-CDS | ATGGCAGAATTCACTGCAGATTT | CAGGGGCCATGACCATGTT |
| GmBEE3-CDS | ATGGCTGAATTCACTGCAGATTT | CAGAGGCCATGGCCATGTT |
| qPCR-GmBEE1a | CAGAACATTGTCCCAGGATG | ATTCTTCTCTCACATACCTGTC |
| qPCR-GmBEE3 | CCGGCTCTTGTTCAATGTGTC | TAGCTTGACCACGTTTGGCT |
| eYFP | GAAGATGCCTCTGCCGACAG | AACTTCAGGGTCAGCTTGCCG |
| GmActin | GGTGGTTCTATCTTGGCATC | CTTTCGCTTCAATAACCCTA |
表 1 用于基因克隆及实时荧光定量PCR扩增的引物序列
Table 1 Primer sequences for gene cloning and real-time fluorescence quantitative PCR amplification
引物名称 Primer name | 上游引物 Forward primer (5′‒3′) | 下游引物 Reverse primer (5′‒3′) |
|---|---|---|
| GmBEE1a-CDS | ATGGCAGAATTCACTGCAGATTT | CAGGGGCCATGACCATGTT |
| GmBEE3-CDS | ATGGCTGAATTCACTGCAGATTT | CAGAGGCCATGGCCATGTT |
| qPCR-GmBEE1a | CAGAACATTGTCCCAGGATG | ATTCTTCTCTCACATACCTGTC |
| qPCR-GmBEE3 | CCGGCTCTTGTTCAATGTGTC | TAGCTTGACCACGTTTGGCT |
| eYFP | GAAGATGCCTCTGCCGACAG | AACTTCAGGGTCAGCTTGCCG |
| GmActin | GGTGGTTCTATCTTGGCATC | CTTTCGCTTCAATAACCCTA |
基因名 Gene name | 基因ID Gene ID | 定位标记 Locus tag | 外显子个数 Number of exons | 内含子个数 Number of introns |
|---|---|---|---|---|
| GmBEE3 | 100777160 | Glyma08g042000 | 6 | 5 |
| GmBEE1a | 100810945 | Glyma05g234500 | 6 | 5 |
| GmBEE1b | 100806942 | Glyma09g183500 | 6 | 5 |
| GmBEE1c | 100807773 | Glyma14g102200 | 6 | 5 |
| GmBEE1d | 100809974 | Glyma17g223100 | 6 | 5 |
| GmBEE1e | 100803700 | Glyma07g092700 | 6 | 5 |
表2 大豆GmBEEs基因的外显子和内含子数量统计
Table 2 Statistics of the number of exons and introns of soybean GmBEEs genes
基因名 Gene name | 基因ID Gene ID | 定位标记 Locus tag | 外显子个数 Number of exons | 内含子个数 Number of introns |
|---|---|---|---|---|
| GmBEE3 | 100777160 | Glyma08g042000 | 6 | 5 |
| GmBEE1a | 100810945 | Glyma05g234500 | 6 | 5 |
| GmBEE1b | 100806942 | Glyma09g183500 | 6 | 5 |
| GmBEE1c | 100807773 | Glyma14g102200 | 6 | 5 |
| GmBEE1d | 100809974 | Glyma17g223100 | 6 | 5 |
| GmBEE1e | 100803700 | Glyma07g092700 | 6 | 5 |
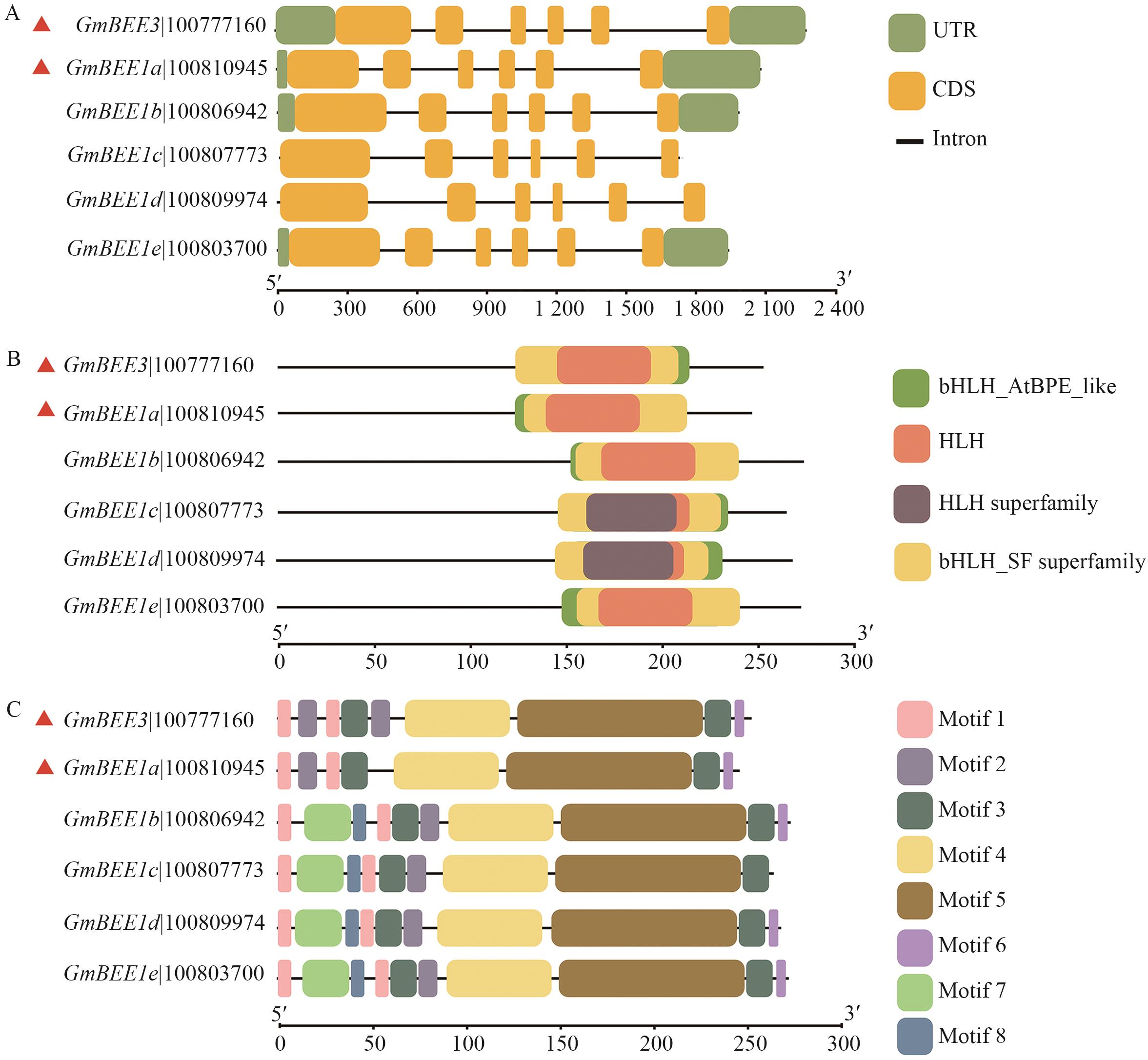
图2 大豆GmBEEs基因结构、基因保守结构域及基序分析A:GmBEEs基因结构; B:GmBEEs保守结构域; C:GmBEEs基序对比
Fig. 2 Analysis of gene structures, gene conserved domains and motifs of soybean GmBEEsA: Structure of GmBEEs gene. B: Conserved domains of GmBEEs. C: Gene motif alignment of GmBEEs
基因名称 Gene name | 基因ID Gene ID | 氨基酸数 Number of amino acids | 分子式 Formula | 分子量 Molecular weight (Da) | 等电点 pI |
|---|---|---|---|---|---|
| GmBEE1a | 100810945 | 246 | C1222H1926N344O381S17 | 28 077.83 | 5.40 |
| GmBEE1b | 100806942 | 273 | C1346H2126N378O416S12 | 30 644.69 | 6.10 |
| GmBEE1c | 100807773 | 264 | C1331H2075N363O408S11 | 30 042.96 | 5.56 |
| GmBEE1d | 100809974 | 268 | C1343H2086N372O419S11 | 30 500.24 | 5.43 |
| GmBEE1e | 100803700 | 272 | C1339H2115N379O413S14 | 30 579.65 | 6.16 |
| GmBEE3 | 100777160 | 252 | C1226H1954N346O387S16 | 28 246.05 | 5.53 |
表3 大豆中5个GmBEE1和1个GmBEE3蛋白理化性质分析
Table 3 Analysis of physicochemical properties of five GmBEE1 and one GmBEE3 protein in soybean
基因名称 Gene name | 基因ID Gene ID | 氨基酸数 Number of amino acids | 分子式 Formula | 分子量 Molecular weight (Da) | 等电点 pI |
|---|---|---|---|---|---|
| GmBEE1a | 100810945 | 246 | C1222H1926N344O381S17 | 28 077.83 | 5.40 |
| GmBEE1b | 100806942 | 273 | C1346H2126N378O416S12 | 30 644.69 | 6.10 |
| GmBEE1c | 100807773 | 264 | C1331H2075N363O408S11 | 30 042.96 | 5.56 |
| GmBEE1d | 100809974 | 268 | C1343H2086N372O419S11 | 30 500.24 | 5.43 |
| GmBEE1e | 100803700 | 272 | C1339H2115N379O413S14 | 30 579.65 | 6.16 |
| GmBEE3 | 100777160 | 252 | C1226H1954N346O387S16 | 28 246.05 | 5.53 |
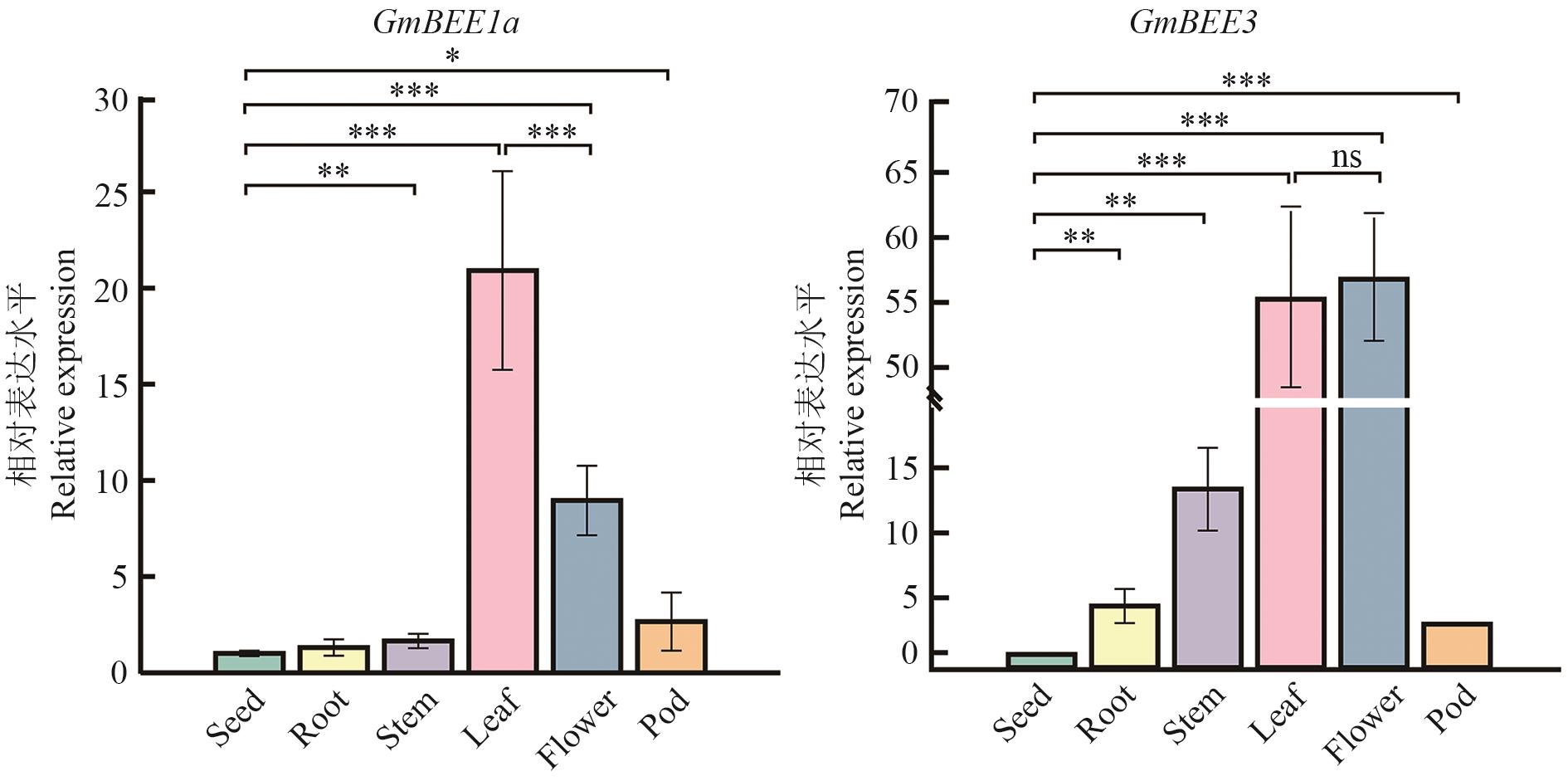
图6 GmBEE1a和GmBEE3组织表达特异性误差线表示平均值±SD;***P<0.001; **P<0.01; *P<0.05;ns表示不存在显著性差异,下同
Fig. 6 Tissue-expression specifity of GmBEE1a and GmBEE3 in soybeanError bars indicate mean ± SD; ***P<0.001; **P<0.01; *P<0.05; ns indicates no significant difference, the same below
| [1] | Liu ZT, Ying H, Chen MY, et al. Optimization of China’s maize and soy production can ensure feed sufficiency at lower nitrogen and carbon footprints [J]. Nat Food, 2021, 2(6): 426-433. |
| [2] | 刘凯, 王欢, 穆月英. 中国大豆进口风险分散及进口来源结构优化—基于替代性与依赖性视角 [J]. 中国油脂, 2025, 50(2): 1-7, 22. |
| Liu K, Wang H, Mu YY. Risk dispersion and optimization of soybean imports for China: Based on substitution and dependence risks [J]. China Oils Fats, 2025, 50(2): 1-7, 22. | |
| [3] | 叶文武, 刘万才, 王源超. 中国大豆病虫害发生现状及全程绿色防控技术研究进展 [J]. 植物保护学报, 2023, 50(2): 265-273. |
| Ye WW, Liu WC, Wang YC. Occurrence status and whole-process green control technologies for soybean diseases and pests in China [J]. J Plant Prot, 2023, 50(2): 265-273. | |
| [4] | 黄新琦, 蔡祖聪. 土壤微生物与作物土传病害控制 [J]. 中国科学院院刊, 2017, 32(6): 593-600. |
| Huang XQ, Cai ZC. Soil microbes and control of soil-borne diseases [J]. Bull Chin Acad Sci, 2017, 32(6): 593-600. | |
| [5] | 杨珍, 戴传超, 王兴祥, 等. 作物土传真菌病害发生的根际微生物机制研究进展 [J]. 土壤学报, 2019, 56(1): 12-22. |
| Yang Z, Dai CC, Wang XX, et al. Advance in research on rhizosphere microbial mechanisms of crop soil-borne fungal diseases [J]. Acta Pedol Sin, 2019, 56(1): 12-22. | |
| [6] | Riechmann JL, Ratcliffe OJ. A genomic perspective on plant transcription factors [J]. Curr Opin Plant Biol, 2000, 3(5): 423-434. |
| [7] | Zhu H, Wang GH, Qian J. Transcription factors as readers and effectors of DNA methylation [J]. Nat Rev Genet, 2016, 17(9): 551-565. |
| [8] | 安昌, 陆琳, 沈梦千, 等. 植物bHLH基因家族研究进展及在药用植物中的应用前景 [J]. 生物技术通报, 2023, 39(10): 1-16. |
| An C, Lu L, Shen MQ, et al. Research progress of bHLH gene family in plants and its application prospects in medical plants [J]. Biotechnol Bull, 2023, 39(10):1-16. | |
| [9] | Wang K, Li MQ, Chang YP, et al. The basic helix-loop-helix transcription factor OsBLR1 regulates leaf angle in rice via brassinosteroid signalling [J]. Plant Mol Biol, 2020, 102(6): 589-602. |
| [10] | Han YC, Hu QF, Gong N, et al. Natural variation in MORE GRAINS 1 regulates grain number and grain weight in rice [J]. J Integr Plant Biol, 2024, 66(7): 1440-1458. |
| [11] | 武家颂, 崔欣茹, 张景荣, 等. 拟南芥bHLH转录因子在根毛发育中的作用 [J]. 杭州师范大学学报: 自然科学版, 2022, 21(2): 144-151. |
| Wu JS, Cui XR, Zhang JR, et al. Role of bHLH transcription factor members in Arabidopsis root hair development [J]. J Hangzhou Norm Univ Nat Sci Ed, 2022, 21(2): 144-151. | |
| [12] | Duan SW, Li MW, Niu YC, et al. A natural non-synonymous single nucleotide polymorphism in GmbHLH113 negates its inhibitory effect on root hair elongation in soybean [J]. Plant J, 2023, 115(3): 742-757. |
| [13] | Song YS, Li SM, Sui Y, et al. SbbHLH85, a bHLH member, modulates resilience to salt stress by regulating root hair growth in sorghum [J]. Theor Appl Genet, 2022, 135(1): 201-216. |
| [14] | Guo WL, Chen BH, Guo YY, et al. Expression of pumpkin CmbHLH87 gene improves powdery mildew resistance in tobacco [J]. Front Plant Sci, 2020, 11: 163. |
| [15] | Cao YY, Liu L, Ma KS, et al. The jasmonate-induced bHLH gene SlJIG functions in terpene biosynthesis and resistance to insects and fungus [J]. J Integr Plant Biol, 2022, 64(5): 1102-1115. |
| [16] | Wang F, Gao YS, Liu YW, et al. BES1-regulated BEE1 controls photoperiodic flowering downstream of blue light signaling pathway in Arabidopsis [J]. New Phytol, 2019, 223(3): 1407-1419. |
| [17] | Poppenberger B, Rozhon W, Khan M, et al. CESTA, a positive regulator of brassinosteroid biosynthesis [J]. EMBO J, 2011, 30(6): 1149-1161. |
| [18] | Chen EY, Wang XQ, Gong Q, et al. A novel GhBEE1-Like gene of cotton causes anther indehiscence in transgenic Arabidopsis under uncontrolled transcription level [J]. Gene, 2017, 627: 49-56. |
| [19] | 颜斌, 武丹阳, 李慧玉. 白桦BpBEE2基因的遗传转化及抗逆性分析 [J]. 植物研究, 2019, 39(2): 287-293. |
| Yan B, Wu DY, Li HY. Genetic transformation and resistance analysis of BpBEE2 gene from Betula platyphylla [J]. Bull Bot Res, 2019, 39(2): 287-293. | |
| [20] | Chang XL, Wei DQ, Zeng YH, et al. Maize-soybean relay strip intercropping reshapes the rhizosphere bacterial community and recruits beneficial bacteria to suppress Fusarium root rot of soybean [J]. Front Microbiol, 2022, 13: 1009689. |
| [21] | Wei DQ, Zhu D, Zhang YF, et al. Characterization of rhizosphere Pseudomonas chlororaphis IRHB3 in the reduction of Fusarium root rot and promotion of soybean growth [J]. Biol Control, 2023, 186: 105349. |
| [22] | Friedrichsen DM, Nemhauser J, Muramitsu T, et al. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth [J]. Genetics, 2002, 162(3): 1445-1456. |
| [23] | 赵婧, 郭茜, 李睿琦, 等. 大豆GmGST基因簇基因序列分析及诱导表达分析 [J]. 生物技术通报, 2025, 41(5): 129-140. |
| Zhao J, Guo Q, Li RQ, et al. Sequence analysis and induced expression analysis of GmGST gene cluster genes in soybean [J]. Biotechnol Bull, 2025, 41(5):129-140. | |
| [24] | Lu R, Li Y, Zhang J, et al. The bHLH/HLH transcription factors GhFP2 and GhACE1 antagonistically regulate fiber elongation in cotton [J]. Plant Physiol, 2022, 189(2): 628-643. |
| [25] | Suzuki H, Seki H, Muranaka T. Insights into the diversification of subclade IVa bHLH transcription factors in Fabaceae [J]. BMC Plant Biol, 2021, 21(1): 109. |
| [26] | Moreno JE, Moreno-Piovano G, Chan RL. The antagonistic basic helix-loop-helix partners BEE and IBH1 contribute to control plant tolerance to abiotic stress [J]. Plant Sci, 2018, 271: 143-150. |
| [27] | Li QF, Lu J, Yu JW, et al. The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multisignal-regulated plant growth [J]. Biochim Biophys Acta BBA Gene Regul Mech, 2018, 1861(6): 561-571. |
| [28] | Liang YG, Yang CY, Ming FY, et al. A bHLH transcription factor, CsSPT, regulates high-temperature resistance in cucumber [J]. Hortic Plant J, 2024, 10(2): 503-514. |
| [29] | Cheng ZH, Song XF, Liu XF, et al. SPATULA and ALCATRAZ confer female sterility and fruit cavity via mediating pistil development in cucumber [J]. Plant Physiol, 2022, 189(3): 1553-1569. |
| [30] | Tang BZ, Liu CY, Li ZQ, et al. Multilayer regulatory landscape during pattern-triggered immunity in rice [J]. Plant Biotechnol J, 2021, 19(12): 2629-2645. |
| [31] | Bai MY, Fan M, Oh E, et al. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis [J]. Plant Cell, 2012, 24(12): 4917-4929. |
| [32] | Malinovsky FG, Batoux M, Schwessinger B, et al. Antagonistic regulation of growth and immunity by the Arabidopsis basichelix-loop-helix transcription factor homolog of brassinosteroid enhanced expression2 interacting with increased leaf inclination1 binding bHLH1[J]. Plant Physiol, 2014, 164(3): 1443-1455. |
| [1] | 张超超, 韩开元, 王彤, 陈仲. 毛白杨PtoYABBY2和PtoYABBY12的克隆及功能分析[J]. 生物技术通报, 2025, 41(9): 256-264. |
| [2] | 史发超, 姜永华, 刘海伦, 文英杰, 严倩. 荔枝LcTFL1基因的克隆与功能分析[J]. 生物技术通报, 2025, 41(9): 159-167. |
| [3] | 黄国栋, 邓宇星, 程宏伟, 但焱南, 周会汶, 吴兰花. 大豆ZIP基因家族鉴定及响应铝胁迫的表达分析[J]. 生物技术通报, 2025, 41(9): 71-81. |
| [4] | 关陟昊, 单治易, 熊赫, 赵瑞雪. 基于计算文献的大豆耦合性状知识发现研究[J]. 生物技术通报, 2025, 41(9): 345-356. |
| [5] | 刘佳丽, 宋经荣, 赵文宇, 张馨元, 赵子洋, 曹一博, 张凌云. 蓝莓R2R3-MYB基因鉴定及类黄酮调控基因表达分析[J]. 生物技术通报, 2025, 41(9): 124-138. |
| [6] | 李玉珍, 李梦丹, 张蔚, 彭婷. 基于月季扩展蛋白基因家族鉴定的野蔷薇RmEXPB2基因功能研究[J]. 生物技术通报, 2025, 41(9): 182-194. |
| [7] | 李亚涛, 张志鹏, 赵梦瑶, 吕镇, 甘恬, 魏浩, 吴书凤, 马玉超. 根瘤菌Bd1的全基因组分析及TetR3对细胞生长和结瘤的负调控功能[J]. 生物技术通报, 2025, 41(9): 289-301. |
| [8] | 黄诗宇, 田姗姗, 杨天为, 高曼熔, 张尚文. 赤苍藤WRI1基因家族的全基因组鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(8): 242-254. |
| [9] | 康琴, 汪霞, 谌明洋, 徐静天, 陈诗兰, 廖平杨, 许文志, 吴卫, 徐东北. 薄荷UV-B受体基因McUVR8的克隆与表达分析[J]. 生物技术通报, 2025, 41(8): 255-266. |
| [10] | 朱丽娟, 张锴, 温晓蕾, 褚佳豪, 史凤玉, 王艳丽. 基于WGCNA挖掘野生大豆耐镉关键基因[J]. 生物技术通报, 2025, 41(8): 124-136. |
| [11] | 翟莹, 计俊杰, 陈炯辛, 于海伟, 李珊珊, 赵艳, 马天意. 异源过表达大豆GmNF-YB24提高转基因烟草抗旱性[J]. 生物技术通报, 2025, 41(8): 137-145. |
| [12] | 牛景萍, 赵婧, 郭茜, 王书宏, 赵晋忠, 杜维俊, 殷丛丛, 岳爱琴. 基于WGCNA鉴定大豆抗大豆花叶病毒NAC转录因子及其诱导表达分析[J]. 生物技术通报, 2025, 41(7): 95-105. |
| [13] | 王芳, 乔帅, 宋伟, 崔鹏娟, 廖安忠, 谭文芳, 杨松涛. 甘薯IbNRT2基因家族全基因组鉴定和表达分析[J]. 生物技术通报, 2025, 41(7): 193-204. |
| [14] | 谭玉荣, 陈东亮, 杨守臻, 赖振光, 唐向民, 孙祖东, 曾维英. 大豆抗豆卷叶螟GmKTI1-like的功能研究[J]. 生物技术通报, 2025, 41(6): 99-108. |
| [15] | 许慧珍, SHANTWANA Ghimire, RAJU Kharel, 岳云, 司怀军, 唐勋. 马铃薯SUMO E3连接酶基因家族分析及StSIZ1基因的克隆与表达模式分析[J]. 生物技术通报, 2025, 41(6): 119-129. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||