








生物技术通报 ›› 2025, Vol. 41 ›› Issue (12): 280-293.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0452
司旭鹏1,2( ), 崔秀文1,2, 欧阳荔枝3, 房丹丹1,2, 裴龙英1,2, 方贵平1,2, 濮希蕾1,2, 马艺沔3, 张争3(
), 崔秀文1,2, 欧阳荔枝3, 房丹丹1,2, 裴龙英1,2, 方贵平1,2, 濮希蕾1,2, 马艺沔3, 张争3( )
)
收稿日期:2025-05-01
出版日期:2025-12-26
发布日期:2026-01-06
通讯作者:
张争,男,博士,研究员,研究方向 :药用植物次生代谢调控;E-mail: zhangzheng@implad.ac.cn作者简介:司旭鹏,男,硕士,讲师,研究方向 :药用植物次生代谢调控;E-mail: sixupeng0527@163.com基金资助:
SI Xu-peng1,2( ), CUI Xiu-wen1,2, OUYANG Li-zhi3, FANG Dan-dan1,2, PEI Long-ying1,2, FANG Gui-ping1,2, PU Xi-lei1,2, MA Yi-mian3, ZHANG Zheng3(
), CUI Xiu-wen1,2, OUYANG Li-zhi3, FANG Dan-dan1,2, PEI Long-ying1,2, FANG Gui-ping1,2, PU Xi-lei1,2, MA Yi-mian3, ZHANG Zheng3( )
)
Received:2025-05-01
Published:2025-12-26
Online:2026-01-06
摘要:
目的 分析新疆阿魏不同部位的转录组信息,发掘倍半萜类成分生物合成关键代谢通路和调控基因,为新疆阿魏分子育种和资源开发提供基因资源。 方法 通过转录组测序获得新疆阿魏根茎叶不同部位的Unigenes序列,并进行生物信息学分析。 结果 经过转录组测序共获得116 517个transcripts,共有44 896条Unigenes序列被注释,新疆阿魏与胡萝卜亚种(Daucus carota subsp. sativus)具有的同源序列匹配度比例最高。19 290个Unigenes被注释于生物过程、细胞组成和分子功能。通过KEGG富集到萜类和聚酮化合物代谢通路(11个通路,204条Unigenes)和其他次生代谢产物合成通路(17个通路,233条Unigenes),进一步研究发现,与倍半萜类合成相关的Unigenes为57个,涉及9个关键酶基因的34条全长转录本。差异表达基因主要富集在MEP途径,6个表达趋势变化明显的酶基因RT-qPCR结果与RNA-seq结果呈现相似的表达模式。表达分析显示倍半萜合成下游关键基因P450和TPS在根中相对表达量最高,DXS和HMGR在叶中的表达量最高,AACT和DXR在茎中表达量最高。同时,转录组共预测到28 017个CDS序列,18 86个转录因子,14 277个SSR位点及9 844对相应引物。 结论 通过转录组分析明确了新疆阿魏倍半萜类物质的生物合成途径,推测根中P450和TPS为倍半萜合成下游关键基因。
司旭鹏, 崔秀文, 欧阳荔枝, 房丹丹, 裴龙英, 方贵平, 濮希蕾, 马艺沔, 张争. 新疆阿魏不同部位转录组特征分析及倍半萜合成相关基因挖掘[J]. 生物技术通报, 2025, 41(12): 280-293.
SI Xu-peng, CUI Xiu-wen, OUYANG Li-zhi, FANG Dan-dan, PEI Long-ying, FANG Gui-ping, PU Xi-lei, MA Yi-mian, ZHANG Zheng. Transcriptome Analysis of Different Parts of Ferula Sinkiangensis K. M. Shen and Exploration of Genes Related to Sesquiterpene Synthesis[J]. Biotechnology Bulletin, 2025, 41(12): 280-293.
基因名 Gene name | 基因ID Gene ID | 引物序列 Primer sequence (5′-3′) |
|---|---|---|
| P450 | Cluster-8529.5884-F | CGGTCCGAGAAGTTATGGGG |
| Cluster-8529.5884-R | TGGCAACCACCCTCTTCATC | |
| DXS | Cluster-8529.15434-F | GCTGGACATGGGTGTATGCT |
| Cluster-8529.15434-R | CTGGAGTTTTGCTCCCCCAT | |
| AACT | Cluster-8529.15178-F | CTGCGGTTAAATCTGCTGGT |
| Cluster-8529.15178-R | GTTACTCCCAAAGGATGCCCC | |
| TPS | Cluster-8529.5582-F | TACTGGATGATGAGGGAGCGA |
| Cluster-8529.5582-R | ACAATACGAGTGGAGCCGC | |
| HMGR | Cluster-8529.7778-F | TTCGAGACGCTTGCACTCAT |
| Cluster-8529.7778-R | TTGCATCCCCAGTGCTACAG | |
| DXR | Cluster-8529.20686-F | TGTCAGAAGCACCAAGACGA |
| Cluster-8529.20686-R | TACAAGAGCGGGACTCAAGC | |
| Actin | Actin-F | TGGTATTGTGCTGGATTCTGGT |
| Actin-R | TGAGATCACCACCAGCAAGG |
表1 RT-qPCR引物序列
Table 1 Primer sequence for RT-qPCR
基因名 Gene name | 基因ID Gene ID | 引物序列 Primer sequence (5′-3′) |
|---|---|---|
| P450 | Cluster-8529.5884-F | CGGTCCGAGAAGTTATGGGG |
| Cluster-8529.5884-R | TGGCAACCACCCTCTTCATC | |
| DXS | Cluster-8529.15434-F | GCTGGACATGGGTGTATGCT |
| Cluster-8529.15434-R | CTGGAGTTTTGCTCCCCCAT | |
| AACT | Cluster-8529.15178-F | CTGCGGTTAAATCTGCTGGT |
| Cluster-8529.15178-R | GTTACTCCCAAAGGATGCCCC | |
| TPS | Cluster-8529.5582-F | TACTGGATGATGAGGGAGCGA |
| Cluster-8529.5582-R | ACAATACGAGTGGAGCCGC | |
| HMGR | Cluster-8529.7778-F | TTCGAGACGCTTGCACTCAT |
| Cluster-8529.7778-R | TTGCATCCCCAGTGCTACAG | |
| DXR | Cluster-8529.20686-F | TGTCAGAAGCACCAAGACGA |
| Cluster-8529.20686-R | TACAAGAGCGGGACTCAAGC | |
| Actin | Actin-F | TGGTATTGTGCTGGATTCTGGT |
| Actin-R | TGAGATCACCACCAGCAAGG |
| 序号 Serial number | 通路ID Pathway ID | 代谢通路 Metabolic pathway | 数量 Quantity |
|---|---|---|---|
| 1 | ko00523 | Polyketide sugar unit biosynthesis | 3 |
| 2 | ko00900 | Terpenoid backbone biosynthesis | 59 |
| 3 | ko00902 | Monoterpenoid biosynthesis | 7 |
| 4 | ko00903 | Limonene degradation | 12 |
| 5 | ko00904 | Diterpenoid biosynthesis | 24 |
| 6 | ko00905 | Brassinosteroid biosynthesis | 18 |
| 7 | ko00906 | Carotenoid biosynthesis | 33 |
| 8 | ko00908 | Zeatin biosynthesis | 34 |
| 9 | ko00909 | Sesquiterpenoid and triterpenoid biosynthesis | 15 |
| 10 | ko01051 | Biosynthesis of ansamycins | 2 |
| 11 | ko01053 | Biosynthesis of siderophore group nonribosomal peptides | 1 |
表2 萜类和聚酮化合物的KEGG通路分析
Table 2 KEGG pathway analysis of terpenoids and polyketides
| 序号 Serial number | 通路ID Pathway ID | 代谢通路 Metabolic pathway | 数量 Quantity |
|---|---|---|---|
| 1 | ko00523 | Polyketide sugar unit biosynthesis | 3 |
| 2 | ko00900 | Terpenoid backbone biosynthesis | 59 |
| 3 | ko00902 | Monoterpenoid biosynthesis | 7 |
| 4 | ko00903 | Limonene degradation | 12 |
| 5 | ko00904 | Diterpenoid biosynthesis | 24 |
| 6 | ko00905 | Brassinosteroid biosynthesis | 18 |
| 7 | ko00906 | Carotenoid biosynthesis | 33 |
| 8 | ko00908 | Zeatin biosynthesis | 34 |
| 9 | ko00909 | Sesquiterpenoid and triterpenoid biosynthesis | 15 |
| 10 | ko01051 | Biosynthesis of ansamycins | 2 |
| 11 | ko01053 | Biosynthesis of siderophore group nonribosomal peptides | 1 |

图4 与药效物质形成密切相关的通路A:其他次生代谢产物合成途径;B:萜类和聚酮化合物的代谢
Fig. 4 Pathway closely related to the formation of pharmacological substancesA: Biosynthesis of other secondary metabolites. B: Metabolism of terpenoids and polyketides
| 骨架 Skeleton | 名称 Name | 数量 Quantity | FPKM (Fragments Per) |
|---|---|---|---|
萜类骨架 Terpenoid skeleton(59)ko00900 | Geranylgeranyl pyrophosphate synthase | 3 | 6.62-14.69 |
| Farnesol kinase | 1 | 4.79-7.41 | |
| 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MDS) | 1 | 5.3-6.11 | |
| 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (MCT) | 3 | 10.67-29.75 | |
| Prenyl protein peptidase | 1 | 3.66-6.47 | |
| Diphosphomevalonate decarboxylase (MVD) | 2 | 4.33-10.76 | |
| Hydroxymethylglutaryl-CoA synthase (HMGCS) | 2 | 6.53-37.72 | |
| Farnesylcysteine lyase | 1 | 39-45.25 | |
| Geranylgeranyl diphosphate reductase | 2 | 3.17-46.74 | |
| Acetyl-CoA C-acetyltransferase (ACAT) | 1 | 23.22-41.51 | |
| Polycis-polyprenyl diphosphate synthase | 2 | 2.29-22.71 | |
| Geranyl diphosphate synthase (GPS) | 3 | 0.39-14.17 | |
| 4-Hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR) | 5 | 0.1-25.06 | |
| Hypothetical protein | 1 | 0.38-1.52 | |
| 1-Deoxy-D-xylulose-5-phosphate synthase (DXS) | 3 | 3.58-43.29 | |
| 3-Hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR) | 3 | 0.43-60.75 | |
| 4-Hydroxy-3-methylbut-2-enyl-diphosphate synthase (HXS) | 1 | 25.93-42.74 | |
| CAAX prenyl protease | 1 | 60.08-65.9 | |
| Mevalonate kinase (MK) | 1 | 16.24-18.02 | |
| Phosphomevalonate kinase (MVK2) | 1 | 8.52-11.37 | |
| Isopentenyl-diphosphate Delta-isomerase | 1 | 75.21-102.72 | |
| Acetyl-CoA C-acetyltransferase | 4 | 0.16-83.75 | |
| Protein-S-isoprenylcysteine O-methyltransferase (STE14) | 1 | 10.42-17.69 | |
| Isopentenyl phosphate kinase | 1 | 8.28-10.05 | |
| Farnesyl diphosphate synthase (FDPS) | 1 | 5.67-7.64 | |
| Dihydroflavonol-4-reductase | 3 | 0.42-18.19 | |
| Prenylcysteine alpha-carboxyl methylesterase | 1 | 3.53-4.22 | |
| Protein farnesyltransferase/geranylgeranyltransferase | 1 | 13.1-14.01 | |
| Geranylgeranyl pyrophosphate synthase/Polyprenyl synthetase | 1 | 6.66-8.04 | |
| 4-Diphosphocytidyl-2-C-methyl-D-erythritol kinase (CMK) | 1 | 12.65-15.84 | |
| Solanesyl-diphosphate synthase | 1 | 2.35-3.24 | |
| Protein farnesyltransferase subunit beta (FNTB) | 1 | 8.47-9.76 | |
| Ditrans,polycis-polyprenyl diphosphate synthase | 1 | 11.54-14.64 | |
| Prenyl protease | 1 | 12.7-13.94 | |
倍半萜及三萜生物合成 Biosynthesis of sesquiterpenes and triterpenes(15)ko00909 | Sesquiterpene synthase | 3 | 0.33-10.01 |
| Beta-amyrin synthase | 2 | 1.05-3.53 | |
| Squalene epoxidase | 4 | 0.14-32.06 | |
| Dihydroflavonol-4-reductase | 3 | 0.16-18.19 | |
| Squalene synthase | 2 | 0.42-58.1 | |
| (3S, 6E)-Nerolidol synthase | 1 | 1.97-3.91 |
表3 新疆阿魏中萜类骨架及倍半萜合成相关的条带
Table 3 Unigenes related to terpenoid backbone and sesquiterpene synthesis in F. sinkiangensis
| 骨架 Skeleton | 名称 Name | 数量 Quantity | FPKM (Fragments Per) |
|---|---|---|---|
萜类骨架 Terpenoid skeleton(59)ko00900 | Geranylgeranyl pyrophosphate synthase | 3 | 6.62-14.69 |
| Farnesol kinase | 1 | 4.79-7.41 | |
| 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MDS) | 1 | 5.3-6.11 | |
| 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (MCT) | 3 | 10.67-29.75 | |
| Prenyl protein peptidase | 1 | 3.66-6.47 | |
| Diphosphomevalonate decarboxylase (MVD) | 2 | 4.33-10.76 | |
| Hydroxymethylglutaryl-CoA synthase (HMGCS) | 2 | 6.53-37.72 | |
| Farnesylcysteine lyase | 1 | 39-45.25 | |
| Geranylgeranyl diphosphate reductase | 2 | 3.17-46.74 | |
| Acetyl-CoA C-acetyltransferase (ACAT) | 1 | 23.22-41.51 | |
| Polycis-polyprenyl diphosphate synthase | 2 | 2.29-22.71 | |
| Geranyl diphosphate synthase (GPS) | 3 | 0.39-14.17 | |
| 4-Hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR) | 5 | 0.1-25.06 | |
| Hypothetical protein | 1 | 0.38-1.52 | |
| 1-Deoxy-D-xylulose-5-phosphate synthase (DXS) | 3 | 3.58-43.29 | |
| 3-Hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR) | 3 | 0.43-60.75 | |
| 4-Hydroxy-3-methylbut-2-enyl-diphosphate synthase (HXS) | 1 | 25.93-42.74 | |
| CAAX prenyl protease | 1 | 60.08-65.9 | |
| Mevalonate kinase (MK) | 1 | 16.24-18.02 | |
| Phosphomevalonate kinase (MVK2) | 1 | 8.52-11.37 | |
| Isopentenyl-diphosphate Delta-isomerase | 1 | 75.21-102.72 | |
| Acetyl-CoA C-acetyltransferase | 4 | 0.16-83.75 | |
| Protein-S-isoprenylcysteine O-methyltransferase (STE14) | 1 | 10.42-17.69 | |
| Isopentenyl phosphate kinase | 1 | 8.28-10.05 | |
| Farnesyl diphosphate synthase (FDPS) | 1 | 5.67-7.64 | |
| Dihydroflavonol-4-reductase | 3 | 0.42-18.19 | |
| Prenylcysteine alpha-carboxyl methylesterase | 1 | 3.53-4.22 | |
| Protein farnesyltransferase/geranylgeranyltransferase | 1 | 13.1-14.01 | |
| Geranylgeranyl pyrophosphate synthase/Polyprenyl synthetase | 1 | 6.66-8.04 | |
| 4-Diphosphocytidyl-2-C-methyl-D-erythritol kinase (CMK) | 1 | 12.65-15.84 | |
| Solanesyl-diphosphate synthase | 1 | 2.35-3.24 | |
| Protein farnesyltransferase subunit beta (FNTB) | 1 | 8.47-9.76 | |
| Ditrans,polycis-polyprenyl diphosphate synthase | 1 | 11.54-14.64 | |
| Prenyl protease | 1 | 12.7-13.94 | |
倍半萜及三萜生物合成 Biosynthesis of sesquiterpenes and triterpenes(15)ko00909 | Sesquiterpene synthase | 3 | 0.33-10.01 |
| Beta-amyrin synthase | 2 | 1.05-3.53 | |
| Squalene epoxidase | 4 | 0.14-32.06 | |
| Dihydroflavonol-4-reductase | 3 | 0.16-18.19 | |
| Squalene synthase | 2 | 0.42-58.1 | |
| (3S, 6E)-Nerolidol synthase | 1 | 1.97-3.91 |
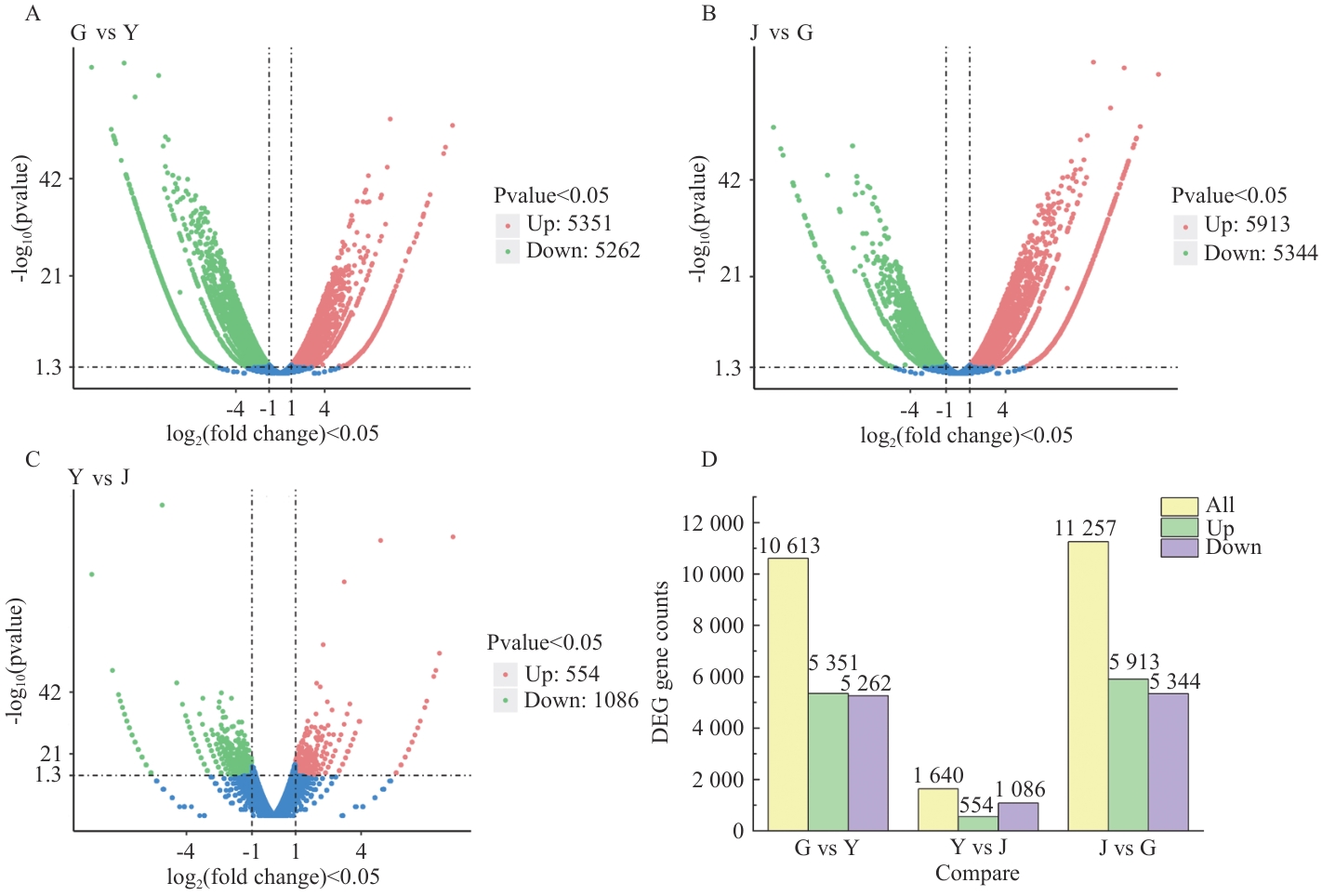
图5 新疆阿魏不同部位差异表达基因分布与表达量A-C:差异表达基因分布;D:差异表达基因的表达量
Fig. 5 Distribution and expression of differentially expressed genes in different parts of F. sinkiangensisA-C: Distribution of differentially expressed genes. D: Expression of differential genes
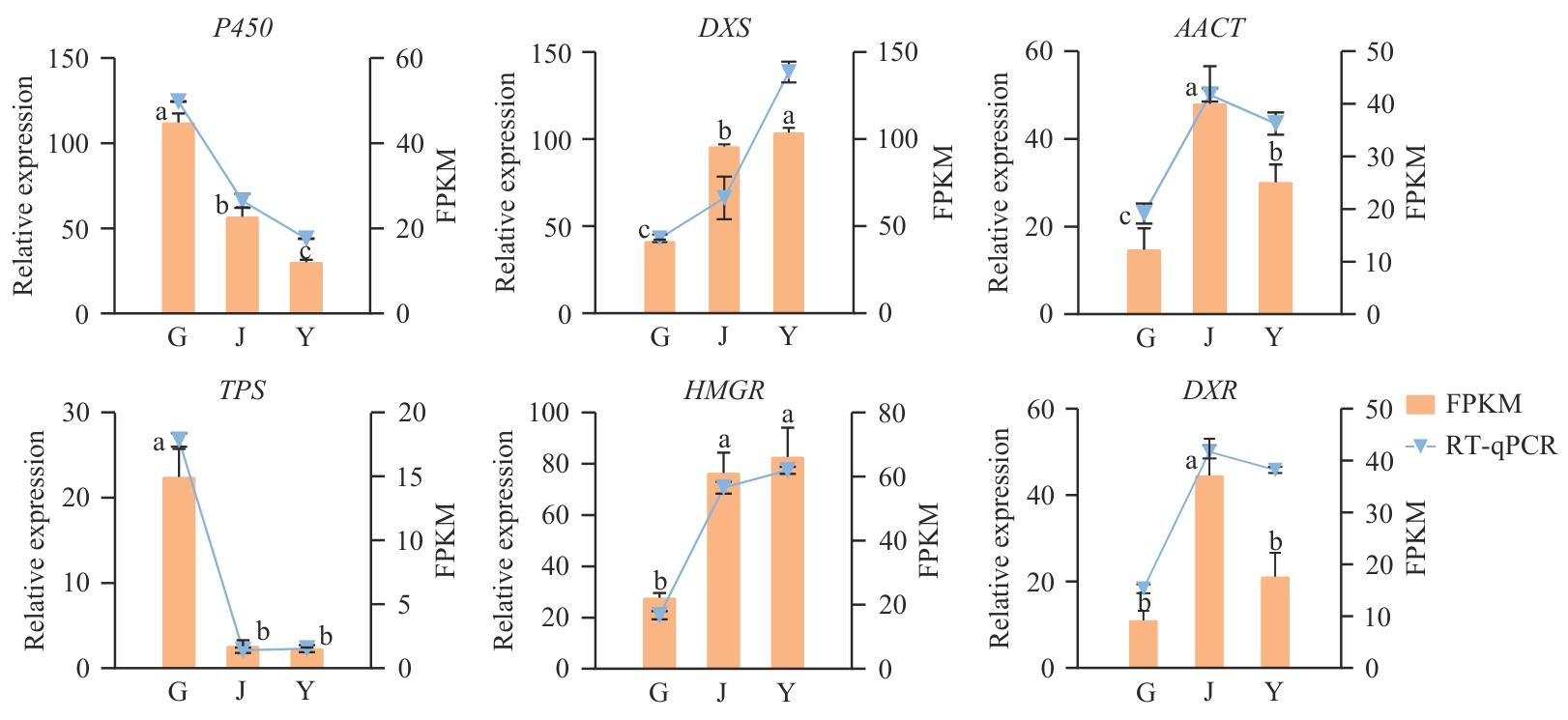
图9 新疆阿魏不同部位倍半萜合成相关基因表达量验证不同小写字母表示0.05水平上差异显著
Fig. 9 Verification of gene expressions related to sesquiterpene synthesis in different parts of F. sinkiangensisData with different lowercase letters indicate significant difference at 0.05 level
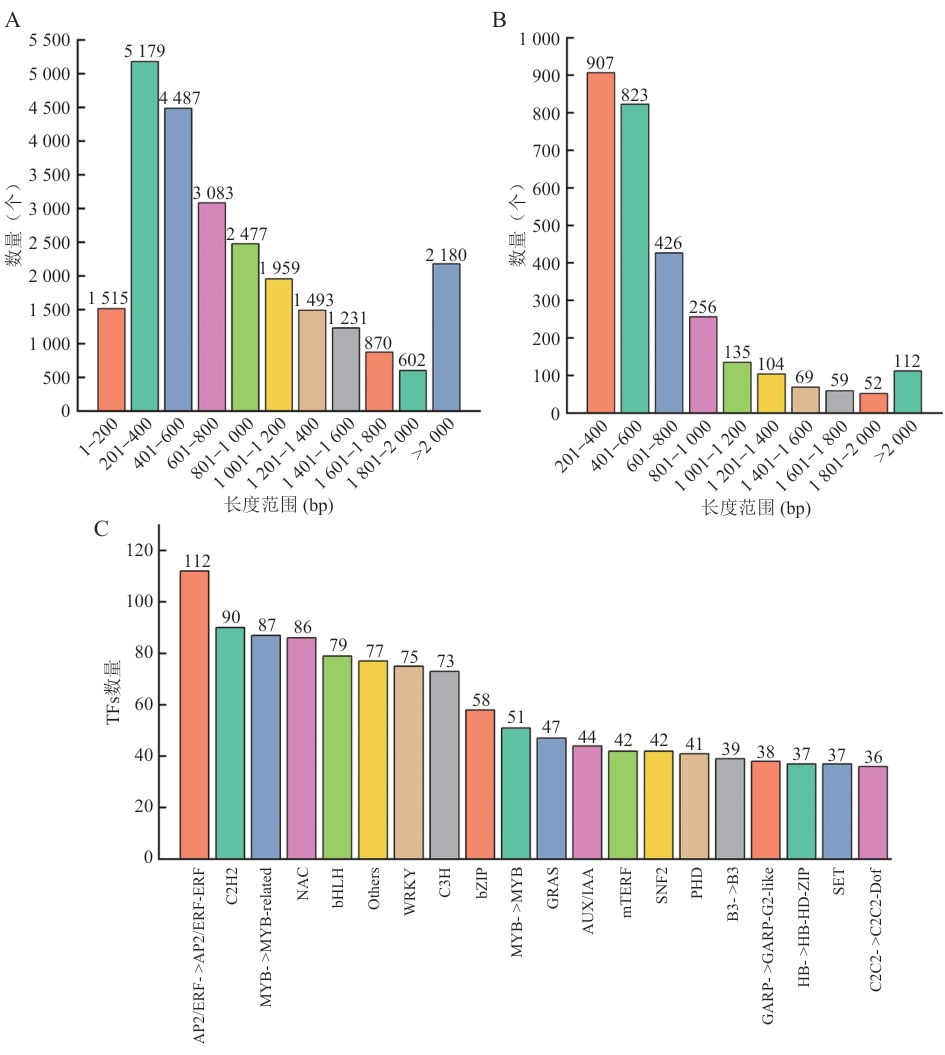
图10 CDS长度分布及转录因子家族的数量A:Blast预测;B:TransDecoder预测;C:TFs数量
Fig. 10 Distribution of CDS lengths and number of transcription factor familyA: Blast prediction. B: TransDecoder prediction. C: Number of TFs
重复基序数目 Number of repeated motifs | SSR位点数目 Number of SSR loci | 总数量 Total quantity | |||||
|---|---|---|---|---|---|---|---|
单核苷酸 Single nucleotide | 二核苷酸 Dinucleotide | 三核苷酸 Trinucleotide | 四核苷酸 Tetranucleotide | 五核苷酸 Pentanucleotide | 六核苷酸 Hexanucleotide | ||
| 5 | 0 | 0 | 844 | 96 | 27 | 41 | 1 008 |
| 6 | 0 | 1800 | 371 | 36 | 8 | 5 | 2 220 |
| 7 | 0 | 1208 | 181 | 18 | 0 | 4 | 1 411 |
| 8 | 0 | 939 | 83 | 5 | 0 | 3 | 1 030 |
| 9 | 0 | 757 | 60 | 1 | 0 | 0 | 818 |
| 10 | 5 | 534 | 42 | 0 | 0 | 0 | 581 |
| 11 | 6 | 338 | 3 | 0 | 0 | 0 | 347 |
| 12 | 7 | 280 | 9 | 1 | 0 | 0 | 297 |
| 13 | 8 | 202 | 9 | 0 | 0 | 0 | 219 |
| 14 | 9 | 227 | 6 | 0 | 0 | 0 | 242 |
| 15 | 15 | 153 | 5 | 1 | 0 | 0 | 174 |
| 16 | 17 | 66 | 1 | 0 | 0 | 0 | 84 |
| 17 | 19 | 72 | 1 | 0 | 0 | 1 | 93 |
| 18 | 21 | 86 | 3 | 0 | 0 | 0 | 110 |
| 19 | 23 | 51 | 2 | 0 | 0 | 0 | 76 |
| 20 | 30 | 54 | 2 | 0 | 0 | 0 | 86 |
| >20 | 198 | 125 | 6 | 0 | 0 | 0 | 329 |
| 总数量 | 358 | 6 892 | 1 628 | 158 | 35 | 54 | 9 125 |
表4 SSR重复基序分布
Table 4 Distribution of SSR repeat motif
重复基序数目 Number of repeated motifs | SSR位点数目 Number of SSR loci | 总数量 Total quantity | |||||
|---|---|---|---|---|---|---|---|
单核苷酸 Single nucleotide | 二核苷酸 Dinucleotide | 三核苷酸 Trinucleotide | 四核苷酸 Tetranucleotide | 五核苷酸 Pentanucleotide | 六核苷酸 Hexanucleotide | ||
| 5 | 0 | 0 | 844 | 96 | 27 | 41 | 1 008 |
| 6 | 0 | 1800 | 371 | 36 | 8 | 5 | 2 220 |
| 7 | 0 | 1208 | 181 | 18 | 0 | 4 | 1 411 |
| 8 | 0 | 939 | 83 | 5 | 0 | 3 | 1 030 |
| 9 | 0 | 757 | 60 | 1 | 0 | 0 | 818 |
| 10 | 5 | 534 | 42 | 0 | 0 | 0 | 581 |
| 11 | 6 | 338 | 3 | 0 | 0 | 0 | 347 |
| 12 | 7 | 280 | 9 | 1 | 0 | 0 | 297 |
| 13 | 8 | 202 | 9 | 0 | 0 | 0 | 219 |
| 14 | 9 | 227 | 6 | 0 | 0 | 0 | 242 |
| 15 | 15 | 153 | 5 | 1 | 0 | 0 | 174 |
| 16 | 17 | 66 | 1 | 0 | 0 | 0 | 84 |
| 17 | 19 | 72 | 1 | 0 | 0 | 1 | 93 |
| 18 | 21 | 86 | 3 | 0 | 0 | 0 | 110 |
| 19 | 23 | 51 | 2 | 0 | 0 | 0 | 76 |
| 20 | 30 | 54 | 2 | 0 | 0 | 0 | 86 |
| >20 | 198 | 125 | 6 | 0 | 0 | 0 | 329 |
| 总数量 | 358 | 6 892 | 1 628 | 158 | 35 | 54 | 9 125 |
| [1] | Yang JR, An Z, Li ZH, et al. Sesquiterpene coumarins from the roots of Ferula sinkiangensis and Ferula teterrima [J]. Chem Pharm Bull, 2006, 54(11): 1595-1598. |
| [2] | Xing YC, Li N, Zhou D, et al. Sesquiterpene coumarins from Ferula sinkiangensis act as neuroinflammation inhibitors [J]. Planta Med, 2017, 83(1/02): 135-142. |
| [3] | Li GZ, Li XJ, Cao L, et al. Steroidal esters from Ferula sinkiangensis [J]. Fitoterapia, 2014, 97: 247-252. |
| [4] | Ye BG, Wang S, Zhang L. Studies on the detoxification effects and acute toxicity of a mixture of Cis-sec-butyl-1-propoenyl disulphide and trans-sec-butyl-1-propoenyl disulphide isolated from crude essential oil of Ferula sinkiangensis K.M. Shen, a Chinese traditional herbal medicine [J]. Nat Prod Res, 2011, 25(12): 1161-1170. |
| [5] | Zhou YT, Xin F, Zhang GQ, et al. Recent advances on bioactive constituents in Ferula [J]. Drug Dev Res, 2017, 78(7): 321-331. |
| [6] | Bao XH, Li YP, Li Q, et al. Racemic norneolignans from the resin of Ferula sinkiangensis and their COX-2 inhibitory activity [J]. Fitoterapia, 2023, 164: 105341. |
| [7] | 李洁, 徐海燕, 贾晓光, 等. 阜康阿魏的化学成分及其生物活性研究进展 [J]. 新疆医科大学学报, 2012, 35(9): 1159-1161. |
| Li J, Xu HY, Jia XG, et al. Research progress on chemical constituents and biological activities of Ferula Fukang [J]. J Xinjiang Med Univ, 2012, 35(9): 1159-1161. | |
| [8] | Wang JC, Zheng Q, Wang HX, et al. Sesquiterpenes and sesquiterpene derivatives from Ferula: their chemical structures, biosynthetic pathways, and biological properties [J]. Antioxidants, 2024, 13(1): 7. |
| [9] | Khayat MT, Alharbi M, Ghazawi KF, et al. Ferula sinkiangensis (Chou-AWei, Chinese Ferula): traditional uses, phytoconstituents, biosynthesis, and pharmacological activities [J]. Plants, 2023, 12(4): 902. |
| [10] | Macrì R, Bava I, Scarano F, et al. In vitro evaluation of ferutinin Rich-Ferula communis L., ssp. glauca, root extract on Doxorubicin-induced cardiotoxicity: antioxidant properties and cell cycle modulation [J]. Int J Mol Sci, 2023, 24(16): 12735. |
| [11] | Mi Y, Jiao K, Xu JK, et al. Kellerin from Ferula sinkiangensis exerts neuroprotective effects after focal cerebral ischemia in rats by inhibiting microglia-mediated inflammatory responses [J]. J Ethnopharmacol, 2021, 269: 113718. |
| [12] | Arjmand Z, Hamburger M, Dastan D. Isolation and purification of terpenoid compounds from Ferula haussknechtii and evaluation of their antibacterial effects [J]. Nat Prod Res, 2023, 37(10): 1617-1624. |
| [13] | Wang JL, Sang CY, Wang J, et al. Sesquiterpene coumarins from Ferula sinkiangensis and their anti-pancreatic cancer effects [J]. Phytochemistry, 2023, 214: 113824. |
| [14] | Li GZ, Wang JC, Li XJ, et al. An unusual sesquiterpene coumarin from the seeds of Ferula sinkiangensis [J]. J Asian Nat Prod Res, 2016, 18(9): 891-896. |
| [15] | 樊丛照. 基于多组学分析的新疆阿魏开花调控机制研究 [D]. 北京: 北京协和医学院, 2024. |
| Fan CZ. Mechanism of flowering regulation in Ferula sinkiangensis based on multi-omies analysis [D]. Beijing: Peking Union Medical College, 2024. | |
| [16] | Tang XH, Li TJ, Hao ZY, et al. Identification of key genes involved in sesquiterpene synthesis in Nardostachys jatamansi based on transcriptome and component analysis [J]. Genes, 2024, 15(12): 1539. |
| [17] | Wang JC, Wang HJ, Zhang M, et al. Sesquiterpene coumarins from Ferula sinkiangensis K.M.Shen and their cytotoxic activities [J]. Phytochemistry, 2020, 180: 112531. |
| [18] | Wang JL, Zhao YM, Qiang Y. Chemical constituents from Ferula sinkiangensis and their chemotaxonomic significance [J]. Biochem Syst Ecol, 2022, 105: 104519. |
| [19] | Ahmed S, Zhan CS, Yang YY, et al. The transcript profile of a traditional Chinese medicine, Atractylodes lancea, revealing its sesquiterpenoid biosynthesis of the major active components [J]. PLoS One, 2016, 11(3): e0151975. |
| [20] | Vranová E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis [J]. Annu Rev Plant Biol, 2013, 64: 665-700. |
| [21] | 陈建, 赵德刚. 植物萜类生物合成相关酶类及其编码基因的研究进展 [J]. 分子植物育种, 2004, 2(6): 757-764. |
| Chen J, Zhao DG. Research advances on the enzymes and their coding gene involved in plant terpene biosynthesis [J]. Mol Plant Breed, 2004, 2(6): 757-764. | |
| [22] | 郭连安, 潘媛, 谭均, 等. 不同生长年限木香转录组分析及倍半萜合成基因挖掘 [J]. 西南农业学报, 2024, 37(9): 2042-2050. |
| Guo LA, Pan Y, Tan J, et al. Transcriptome analysis of Aucklandiae lappa from different growth years and mining genes related to sesquiterpenes synthesis [J]. Southwest China J Agric Sci, 2024, 37(9): 2042-2050. | |
| [23] | Kim BR, Kim SU, Chang YJ. Differential expression of three 1-deoxy-D-xylulose-5-phosphate synthase genes in rice [J]. Biotechnol Lett, 2005, 27(14): 997-1001. |
| [24] | 曾珊珊, 高婷, 谷梦雅, 等. 基于黄棪(Camellia sinensis)基因组的DXS基因家族鉴定及表达 [J]. 应用与环境生物学报, 2024, 30(4): 811-817. |
| Zeng SS, Gao T, Gu MY, et al. Genome-wide identification and expression of the DXS gene family in Huangdan (Camellia sinensis) [J]. Chin J Appl Environ Biol, 2024, 30(4): 811-817. | |
| [25] | Zhang XW, Guo JW, Cheng FY, et al. Cytochrome P450 enzymes in fungal natural product biosynthesis [J]. Nat Prod Rep, 2021, 38(6): 1072-1099. |
| [26] | 高世玺, 戎梅, 彭俊祥, 等. 细胞色素P450在植物倍半萜类化合物生物合成中的作用研究进展 [J]. 药学学报, 2024, 59(2): 313-321. |
| Gao SX, Rong M, Peng JX, et al. Research progress on the role of cytochrome P450 in plant sesquiterpene biosynthesis [J]. Acta Pharm Sin, 2024, 59(2): 313-321. | |
| [27] | Chen DH, Liu CJ, Ye HC, et al. Ri-mediated transformation of Artemisia annua with a recombinant farnesyl diphosphate synthase gene for artemisinin production [J]. Plant Cell Tissue Organ Cult, 1999, 57(3): 157-162. |
| [28] | Ichinose H, Kitaoka T. Insight into metabolic diversity of the brown-rot basidiomycete Postia placenta responsible for sesquiterpene biosynthesis: semi-comprehensive screening of cytochrome P450 monooxygenase involved in protoilludene metabolism [J]. Microb Biotechnol, 2018, 11(5): 952-965. |
| [29] | Shoji T, Yuan L. ERF gene clusters: working together to regulate metabolism [J]. Trends Plant Sci, 2021, 26(1): 23-32. |
| [30] | Liu YH, Khan AR, Gan YB. C2H2 zinc finger proteins response to abiotic stress in plants [J]. Int J Mol Sci, 2022, 23(5): 2730. |
| [31] | 李超颖. 新疆阿魏化学成分及高效液相指纹图谱研究 [D]. 乌鲁木齐: 新疆大学, 2014. |
| Li CY. Studies of the chemical constituents and the HPLC fingerprints of Ferula sinkiangensis K.M.Shen [D]. Urumqi: Xinjiang University, 2014. | |
| [32] | 田再民, 张立平, 孙庆林, 等. 小麦数量性状基因定位研究进展 [J]. 内蒙古农业科技, 2007, 35(3): 68-71. |
| Tian ZM, Zhang LP, Sun QL, et al. Research review on wheat QTLs [J]. Inn Mong Agric Sci Technol, 2007, 35(3): 68-71. | |
| [33] | Fan CJ, Liu QY, Zeng BS, et al. Development of simple sequence repeat (SSR) markers and genetic diversity analysis in blackwood (Acacia melanoxylon) clones in China [J]. Silvae Genet, 2016, 65(1): 49-54. |
| [34] | Cregan PB, Akkaya MS, Bhagwat AA, et al. Length polymorphisms of simple sequence repeat (SSR) DNA as molecular markers in plants [M]// United Kingdom: CRC Press, Plant genome analysis, 1994: 47-56. |
| [35] | Kaur S, Pembleton LW, Cogan NO, et al. Transcriptome sequencing of field pea and faba bean for discovery and validation of SSR genetic markers [J]. BMC Genom, 2012, 13(1): 104. |
| [36] | Yang QW, Jiang YJ, Wang YP, et al. SSR loci analysis in transcriptome and molecular marker development in Polygonatum sibiricum . [J]. BioMed Res Int, 2022, 2022(1): 4237913. |
| [37] | Liu YL, Zhang PF, Song ML, et al. Transcriptome analysis and development of SSR molecular markers in Glycyrrhiza uralensis fisch [J]. PLoS One, 2015, 10(11): e0143017. |
| [1] | 刘建国, 刘格儿, 郭颖欣, 王斌, 王玉昆, 卢金凤, 黄文庭, 朱云娜. 转录组和代谢组联合解析‘桂柚1号’和‘沙田柚’果实品质差异[J]. 生物技术通报, 2025, 41(9): 168-181. |
| [2] | 刘泽洲, 段乃彬, 岳丽昕, 王清华, 姚行浩, 高莉敏, 孔素萍. 大蒜叶片蜡质成分分析及蜡质缺失基因Ggl-1筛选[J]. 生物技术通报, 2025, 41(9): 219-231. |
| [3] | 闫梦阳, 梁晓阳, 戴君昂, 张妍, 关团, 张辉, 刘良波, 孙志华. 阿莫西林降解菌的筛选及降解机制研究[J]. 生物技术通报, 2025, 41(9): 314-325. |
| [4] | 卢瑶, 袁平平, 金鑫, 毛向红, 范向斌, 白小东. 基于SSR标记的马铃薯野生种和地方种遗传多样性分析和指纹图谱构建[J]. 生物技术通报, 2025, 41(9): 94-104. |
| [5] | 白雨果, 李婉迪, 梁建萍, 石志勇, 卢庚龙, 刘红军, 牛景萍. 哈茨木霉T9131对黄芪幼苗的促生机理[J]. 生物技术通报, 2025, 41(8): 175-185. |
| [6] | 柴军发, 洪波, 贾彦霞. 转录组和代谢组联合分析三株蜡蚧轮枝菌菌株毒力差异[J]. 生物技术通报, 2025, 41(8): 311-321. |
| [7] | 裴红霞, 汪露瑶, 李生梅, 高晶霞. 基于SCoT、SRAP和SSR分子标记的220份辣椒种质资源遗传多样性分析[J]. 生物技术通报, 2025, 41(8): 165-174. |
| [8] | 王月琛, 韩鑫骐, 魏文敏, 崔兆兰, 罗阳美, 陈鹏如, 王海岗, 刘龙龙, 张莉, 王纶. 黍稷落粒的生物学基础研究及落粒调控基因的鉴定[J]. 生物技术通报, 2025, 41(7): 164-171. |
| [9] | 张越, 毕钰, 慕雪男, 郑子薇, 王志刚, 徐伟慧. 小麦赤霉病拮抗菌JB7的生防特性[J]. 生物技术通报, 2025, 41(7): 261-271. |
| [10] | 段敏杰, 李怡斐, 王春萍, 黄任中, 黄启中, 张世才. 辣椒果实颜色性状与SSR分子标记的关联分析及指纹图谱构建[J]. 生物技术通报, 2025, 41(7): 81-94. |
| [11] | 李成花, 豆飞飞, 任毓昭, 刘彩霞, 刘凤楼, 王掌军, 李清峰. 外施水杨酸对白粉菌侵染小麦的影响及白粉菌转录组分析[J]. 生物技术通报, 2025, 41(7): 272-280. |
| [12] | 郭秀娟, 冯宇, 吴瑞香, 王利琴, 杨建春. Ca2+处理对胡麻种子萌发影响的转录组分析[J]. 生物技术通报, 2025, 41(7): 139-149. |
| [13] | 段永红, 杨欣, 于冠群, 夏俊俊, 宋陆帅, 白小东, 彭锁堂. 125份马铃薯种质资源遗传多样性及主成分分析[J]. 生物技术通报, 2025, 41(6): 130-143. |
| [14] | 胡若群, 曾菁菁, 梁婉凤, 曹佳玉, 黄小苇, 梁晓英, 仇明月, 陈莹. 转录组和代谢组联合分析探究不同遮光条件下金线莲类胡萝卜素合成代谢机制[J]. 生物技术通报, 2025, 41(5): 231-243. |
| [15] | 唐游, 赵俊伟, 孙兰茜, 李翔. 多组学技术在植物代谢通路解析中的联合应用[J]. 生物技术通报, 2025, 41(4): 76-87. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||