








生物技术通报 ›› 2025, Vol. 41 ›› Issue (5): 231-243.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1007
• 研究报告 • 上一篇
胡若群( ), 曾菁菁, 梁婉凤, 曹佳玉, 黄小苇, 梁晓英, 仇明月, 陈莹(
), 曾菁菁, 梁婉凤, 曹佳玉, 黄小苇, 梁晓英, 仇明月, 陈莹( )
)
收稿日期:2024-10-15
出版日期:2025-05-26
发布日期:2025-06-05
通讯作者:
陈莹,女,博士,副教授,研究方向 :观赏药用植物和园林生态;E-mail: 000q020057@fafu.edu.cn作者简介:胡若群,女,硕士研究生,研究方向 :观赏药用植物和园林生态;E-mail: 357031900@qq.com
基金资助:
HU Ruo-qun( ), ZENG Jing-jing, LIANG Wan-feng, CAO Jia-yu, HUANG Xiao-wei, LIANG Xiao-ying, QIU Ming-yue, CHEN Ying(
), ZENG Jing-jing, LIANG Wan-feng, CAO Jia-yu, HUANG Xiao-wei, LIANG Xiao-ying, QIU Ming-yue, CHEN Ying( )
)
Received:2024-10-15
Published:2025-05-26
Online:2025-06-05
摘要:
目的 探究不同遮光条件对金线莲类胡萝卜素生物合成代谢的影响,解析其类胡萝卜素积累分子机制。 方法 以金线莲为实验材料,分别选取不同遮光处理(T1:遮光25%,T2:遮光50%,T3:遮光75%)叶片,采用高通量RNA测序技术(RNA-Seq)和液相色谱-质谱联用技术(LC-MS/MS)获得金线莲的转录组学与代谢组学数据。通过生物信息学分析,鉴定和量化差异基因与代谢物,并探讨它们之间的关系。 结果 在不同遮光条件下,一共鉴定出20个差异表达基因与21种差异代谢物,其中ZEP、LUT5、LUT1、LCYE、NCED1等关键酶基因显著表达,调控玉米黄质、新黄质、金盏花黄质等类胡萝卜素代谢物的变化,并在75%的遮光处理下含量最高。同时预测到22个转录因子参与调控包括ZEP、ZDS、CYP707A等在内的14个差异表达基因,其中有15个转录因子响应脱落酸、赤霉素等5类植物激素元件参与调控类胡萝卜素生物合成;并挑选8个参与类胡萝卜素生物合成的差异表达基因进行RT-qPCR验证,结果表明8个基因的表达趋势与测序结果一致。 结论 不同遮光条件下金线莲叶片中共有15个转录因子响应5类植物激素元件,进而调控ZEP、NCED1、CCD7等差异基因的表达,使玉米黄质、新黄质等与类胡萝卜素相关的代谢物在75%的遮光处理下显著积累。
胡若群, 曾菁菁, 梁婉凤, 曹佳玉, 黄小苇, 梁晓英, 仇明月, 陈莹. 转录组和代谢组联合分析探究不同遮光条件下金线莲类胡萝卜素合成代谢机制[J]. 生物技术通报, 2025, 41(5): 231-243.
HU Ruo-qun, ZENG Jing-jing, LIANG Wan-feng, CAO Jia-yu, HUANG Xiao-wei, LIANG Xiao-ying, QIU Ming-yue, CHEN Ying. Integrated Transcriptome and Metabolome Analysis to Explore the Carotenoid Synthesis and Metabolism Mechanism in Anoectochilus roxburghii under Different Shading Conditions[J]. Biotechnology Bulletin, 2025, 41(5): 231-243.

图1 不同遮光处理的金线莲T1:遮光率25%;T2:遮光率50%;T3:遮光率75%。下同
Fig. 1 Anoectochilus roxburghii under different shading treatmentsT1: 25% shading treatment; T2: 50% shading treatment; T3: 75% shading treatment. The same below
| 基因名称Gene name | 基因ID Gene ID | 正向引物 Forward primer (5′-3′) | 反向引物 Revese primer(5′-3′) |
|---|---|---|---|
| Actin | Reference gene | AGCATGAAGATTAAGGTGGTTG | CTGAGAGAAGCAAGGATGGA |
| CYP707A | TRINITY_DN8883_c0_g1 | TCCAGCGCGGAGAATATCAC | GTGTTGATGAGGCGGCTTTC |
| CCD7 | TRINITY_DN10188_c0_g1 | CGATGCTGAAAGAACAGCCG | CCAAAGGCAGAACAACCTGC |
| NCED1 | TRINITY_DN9311_c2_g1 | TTCAGACTACTCGCCGCTTC | TGGTTCAGGCCGTCTTTCTC |
| LUT5 | TRINITY_DN2482_c0_g1 | CCTGTTGAAAGCTCAGAGGC | ATGCAGAATGGAACCCAGGTG |
| LUT1 | TRINITY_DN1083_c0_g1 | ATCCCAGTGGCTAATGCTCG | TGATGACGACGAAGTCACGG |
| ZEP | TRINITY_DN5360_c0_g1 | GGCGACGAGCTCAGGAATAA | CTCAACTGCACGGACTTCCT |
| AOG | TRINITY_DN513_c0_g1 | TCGGCTATGCCATGTATCCG | TGATATGCCTCTGAACCGCC |
| ZDS | TRINITY_DN1558_c1_g1 | GCAGGACTACATCGACAGCA | CTCATTGGCTAGTGCCTCGT |
表1 RT-qPCR实验所用引物列表
Table 1 Primers for RT-qPCR
| 基因名称Gene name | 基因ID Gene ID | 正向引物 Forward primer (5′-3′) | 反向引物 Revese primer(5′-3′) |
|---|---|---|---|
| Actin | Reference gene | AGCATGAAGATTAAGGTGGTTG | CTGAGAGAAGCAAGGATGGA |
| CYP707A | TRINITY_DN8883_c0_g1 | TCCAGCGCGGAGAATATCAC | GTGTTGATGAGGCGGCTTTC |
| CCD7 | TRINITY_DN10188_c0_g1 | CGATGCTGAAAGAACAGCCG | CCAAAGGCAGAACAACCTGC |
| NCED1 | TRINITY_DN9311_c2_g1 | TTCAGACTACTCGCCGCTTC | TGGTTCAGGCCGTCTTTCTC |
| LUT5 | TRINITY_DN2482_c0_g1 | CCTGTTGAAAGCTCAGAGGC | ATGCAGAATGGAACCCAGGTG |
| LUT1 | TRINITY_DN1083_c0_g1 | ATCCCAGTGGCTAATGCTCG | TGATGACGACGAAGTCACGG |
| ZEP | TRINITY_DN5360_c0_g1 | GGCGACGAGCTCAGGAATAA | CTCAACTGCACGGACTTCCT |
| AOG | TRINITY_DN513_c0_g1 | TCGGCTATGCCATGTATCCG | TGATATGCCTCTGAACCGCC |
| ZDS | TRINITY_DN1558_c1_g1 | GCAGGACTACATCGACAGCA | CTCATTGGCTAGTGCCTCGT |
| 代谢物 Metabolite | 分子式 Molecular formula | KEGG注释编号 KEGG annotation number |
|---|---|---|
| 二氢菜豆酸 Dihydrophaseic acid | C15H22O5 | C15971 |
| 植物预黄质二磷酸 Prephytoene diphosphate | C40H68O7P2 | C03427 |
| 黄质醛酸 Xanthoxic acid | C15H22O4 | C13454 |
| 圆酵母素Torulene | C40H54 | C08613 |
| 念珠藻黄素 Nostoxanthin | C40H56O4 | C16284 |
| 羟基氯菌烯葡萄糖苷 Hydroxychlorobactene glucoside | C46H64O6 | C15914 |
| 环氧玉米黄质 Antheraxanthin | C40H56O3 | C08579 |
| 辣椒玉红素 Capsorubin | C40H56O4 | C08585 |
| 角黄素 Canthaxanthin | C40H52O2 | C08583 |
| 金盏花黄质 Adonixanthin | C40H54O3 | C15968 |
| 新黄质 Neoxanthin | C40H56O4 | C08606 |
9-顺式-10'-Apo-β-胡萝卜素 9-cis-10'-Apo-beta-Carotenal | C27H36O | C20692 |
玉米黄质二葡萄糖苷 Zeaxanthin diglucoside | C52H76O12 | C15969 |
脱落酸葡萄糖酯 Abscisic acid glucose ester | C21H30O9 | C15970 |
独脚金内酯 ABC-环 Strigolactone ABC-rings | C14H18O3 | C18036 |
| 4,4'-二聚苯二醛 4,4'-Diapolycopenedial | C30H36O2 | C19798 |
| 葡萄球菌黄质 Staphyloxanthin | C51H78O8 | C16148 |
| 羟基氯菌烯 Hydroxychlorobactene | C40H54O | C15911 |
| 玉米黄质 Zeaxanthin | C40H56O2 | C06098 |
| 3-羟基海胆酮 3-Hydroxyechinenone | C40H54O2 | C15966 |
| (3S,2'S)-4-酮米索2'-α-L-岩藻糖苷 (3S,2'S)-4-Ketomyxol 2'-alpha-L-fucoside | C46H64O8 | C15942 |
表2 不同遮光处理下金线莲叶片中类胡萝卜素代谢物
Table 2 Carotenoid metabolites in A. roxburghii leaves under different shading treatments
| 代谢物 Metabolite | 分子式 Molecular formula | KEGG注释编号 KEGG annotation number |
|---|---|---|
| 二氢菜豆酸 Dihydrophaseic acid | C15H22O5 | C15971 |
| 植物预黄质二磷酸 Prephytoene diphosphate | C40H68O7P2 | C03427 |
| 黄质醛酸 Xanthoxic acid | C15H22O4 | C13454 |
| 圆酵母素Torulene | C40H54 | C08613 |
| 念珠藻黄素 Nostoxanthin | C40H56O4 | C16284 |
| 羟基氯菌烯葡萄糖苷 Hydroxychlorobactene glucoside | C46H64O6 | C15914 |
| 环氧玉米黄质 Antheraxanthin | C40H56O3 | C08579 |
| 辣椒玉红素 Capsorubin | C40H56O4 | C08585 |
| 角黄素 Canthaxanthin | C40H52O2 | C08583 |
| 金盏花黄质 Adonixanthin | C40H54O3 | C15968 |
| 新黄质 Neoxanthin | C40H56O4 | C08606 |
9-顺式-10'-Apo-β-胡萝卜素 9-cis-10'-Apo-beta-Carotenal | C27H36O | C20692 |
玉米黄质二葡萄糖苷 Zeaxanthin diglucoside | C52H76O12 | C15969 |
脱落酸葡萄糖酯 Abscisic acid glucose ester | C21H30O9 | C15970 |
独脚金内酯 ABC-环 Strigolactone ABC-rings | C14H18O3 | C18036 |
| 4,4'-二聚苯二醛 4,4'-Diapolycopenedial | C30H36O2 | C19798 |
| 葡萄球菌黄质 Staphyloxanthin | C51H78O8 | C16148 |
| 羟基氯菌烯 Hydroxychlorobactene | C40H54O | C15911 |
| 玉米黄质 Zeaxanthin | C40H56O2 | C06098 |
| 3-羟基海胆酮 3-Hydroxyechinenone | C40H54O2 | C15966 |
| (3S,2'S)-4-酮米索2'-α-L-岩藻糖苷 (3S,2'S)-4-Ketomyxol 2'-alpha-L-fucoside | C46H64O8 | C15942 |
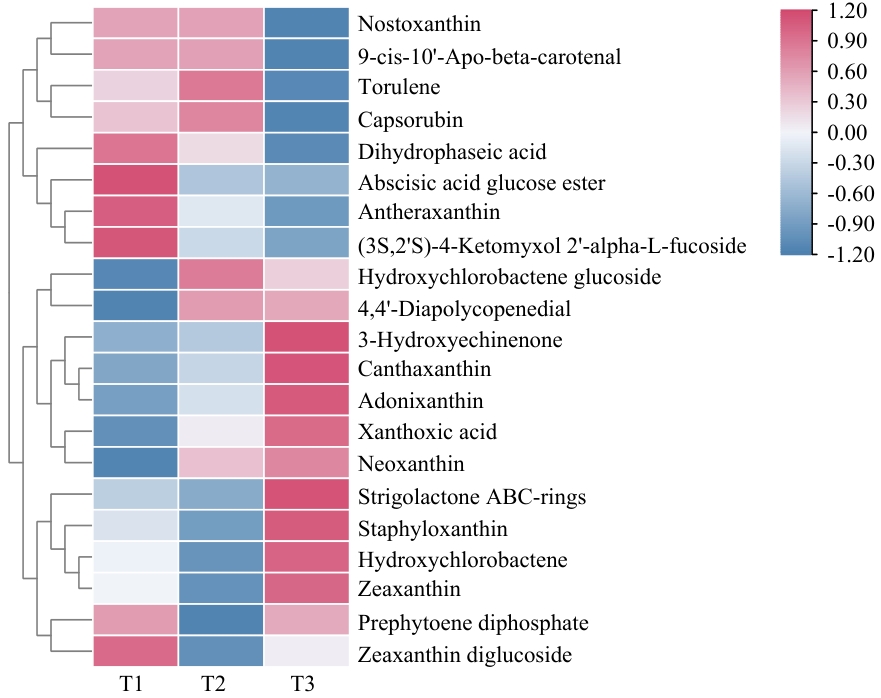
图2 不同遮光处理下金线莲叶片中差异类胡萝卜素代谢物热图粉色代表上调,蓝色代表下调。下同
Fig. 2 Heatmap of differential carotenoid metabolites in A. roxburghii leaves under different shading treatmentsPink indicate up-regulation, blue indicates down-regulation. The same below
| 基因 Gene | 缩写 Abbreviation | 基因ID Gene ID | 酶编号 EC number | KO号 Enzyme KO |
|---|---|---|---|---|
| (+)-脱落酸8'-羟化酶 (+)-abscisic acid 8'-hydroxylase | CYP707A | TRINITY_DN13073_c0_g1 | [EC:1.14.14.137] | K09843 |
| TRINITY_DN1970_c0_g1 | [EC:1.14.14.137] | K09843 | ||
| TRINITY_DN8883_c0_g2 | [EC:1.14.14.137] | K09843 | ||
| TRINITY_DN25069_c0_g1 | [EC:1.14.14.137] | K09843 | ||
| TRINITY_DN5389_c0_g1 | [EC:1.14.14.137] | K09843 | ||
| TRINITY_DN8883_c0_g1 | [EC:1.14.14.137] | K09843 | ||
| 玉米黄质环氧化酶 Zeaxanthin epoxidase | ZEP | TRINITY_DN5360_c0_g1 | [EC:1.14.15.21] | K09838 |
| TRINITY_DN11443_c0_g1 | [EC:1.14.15.21] | K09838 | ||
9-顺式-β-胡萝卜素9',10'-裂解双加氧酶 10-9-cis-beta-carotene 9',10'-cleaving dioxygenase | CCD7 | TRINITY_DN10188_c0_g1 | [EC:1.13.11.68] | K17912 |
| TRINITY_DN8303_c0_g1 | [EC:1.13.11.68] | K17912 | ||
| β-胡萝卜素异构酶 Beta-carotene isomerase | DWARF27 | TRINITY_DN5387_c0_g1 | [EC:5.2.1.14] | K17911 |
| TRINITY_DN840_c0_g1 | [EC:5.2.1.14] | K17911 | ||
9-顺式环氧类胡萝卜素双加氧酶 9-cis-epoxycarotenoid dioxygenase | NCED1 | TRINITY_DN1390_c0_g1 | [EC:1.13.11.51] | K09840 |
| TRINITY_DN562_c3_g1 | [EC:1.13.11.51] | K09840 | ||
| TRINITY_DN9311_c2_g1 | [EC:1.13.11.51] | K09840 | ||
| ε-胡萝卜素羟化酶 Carotenoid epsilon hydroxylase | LUT1 | TRINITY_DN1083_c0_g1 | [EC:1.14.14.158] | K09837 |
| β-胡萝卜素环羟化酶 Beta-ring hydroxylase | LUT5 | TRINITY_DN2482_c0_g1 | [EC:1.14.-.-] | K15747 |
| 胡萝卜素去饱和酶 Zeta-carotene desaturase | ZDS | TRINITY_DN1558_c1_g1 | [EC:1.3.5.6] | K00514 |
| 番茄红素ε环化酶 Lycopene epsilon-cyclase | LCYE | TRINITY_DN29073_c0_g1 | [EC:5.5.1.18] | K06444 |
| 脱落酸β-葡萄糖基转移酶 Abscisate beta-glucosyltransferase | AOG | TRINITY_DN513_c0_g1 | [EC:2.4.1.263] | K14595 |
表3 不同遮光下金线莲叶片中类胡萝卜素差异表达基因
Table 3 Differential expressions of carotenoid-related genes in A. roxburghii leaves under different shading conditions
| 基因 Gene | 缩写 Abbreviation | 基因ID Gene ID | 酶编号 EC number | KO号 Enzyme KO |
|---|---|---|---|---|
| (+)-脱落酸8'-羟化酶 (+)-abscisic acid 8'-hydroxylase | CYP707A | TRINITY_DN13073_c0_g1 | [EC:1.14.14.137] | K09843 |
| TRINITY_DN1970_c0_g1 | [EC:1.14.14.137] | K09843 | ||
| TRINITY_DN8883_c0_g2 | [EC:1.14.14.137] | K09843 | ||
| TRINITY_DN25069_c0_g1 | [EC:1.14.14.137] | K09843 | ||
| TRINITY_DN5389_c0_g1 | [EC:1.14.14.137] | K09843 | ||
| TRINITY_DN8883_c0_g1 | [EC:1.14.14.137] | K09843 | ||
| 玉米黄质环氧化酶 Zeaxanthin epoxidase | ZEP | TRINITY_DN5360_c0_g1 | [EC:1.14.15.21] | K09838 |
| TRINITY_DN11443_c0_g1 | [EC:1.14.15.21] | K09838 | ||
9-顺式-β-胡萝卜素9',10'-裂解双加氧酶 10-9-cis-beta-carotene 9',10'-cleaving dioxygenase | CCD7 | TRINITY_DN10188_c0_g1 | [EC:1.13.11.68] | K17912 |
| TRINITY_DN8303_c0_g1 | [EC:1.13.11.68] | K17912 | ||
| β-胡萝卜素异构酶 Beta-carotene isomerase | DWARF27 | TRINITY_DN5387_c0_g1 | [EC:5.2.1.14] | K17911 |
| TRINITY_DN840_c0_g1 | [EC:5.2.1.14] | K17911 | ||
9-顺式环氧类胡萝卜素双加氧酶 9-cis-epoxycarotenoid dioxygenase | NCED1 | TRINITY_DN1390_c0_g1 | [EC:1.13.11.51] | K09840 |
| TRINITY_DN562_c3_g1 | [EC:1.13.11.51] | K09840 | ||
| TRINITY_DN9311_c2_g1 | [EC:1.13.11.51] | K09840 | ||
| ε-胡萝卜素羟化酶 Carotenoid epsilon hydroxylase | LUT1 | TRINITY_DN1083_c0_g1 | [EC:1.14.14.158] | K09837 |
| β-胡萝卜素环羟化酶 Beta-ring hydroxylase | LUT5 | TRINITY_DN2482_c0_g1 | [EC:1.14.-.-] | K15747 |
| 胡萝卜素去饱和酶 Zeta-carotene desaturase | ZDS | TRINITY_DN1558_c1_g1 | [EC:1.3.5.6] | K00514 |
| 番茄红素ε环化酶 Lycopene epsilon-cyclase | LCYE | TRINITY_DN29073_c0_g1 | [EC:5.5.1.18] | K06444 |
| 脱落酸β-葡萄糖基转移酶 Abscisate beta-glucosyltransferase | AOG | TRINITY_DN513_c0_g1 | [EC:2.4.1.263] | K14595 |

图3 不同遮光处理下金线莲叶片中类胡萝卜素相关差异表达基因热图粉色代表上调,蓝色代表下调。下同
Fig. 3 Heatmap of carotenoid-related differentially expressed genes in A. roxburghii leaves under different shading treatmentsPink indicates up-regulation, blue indicates down-regulation. The same below
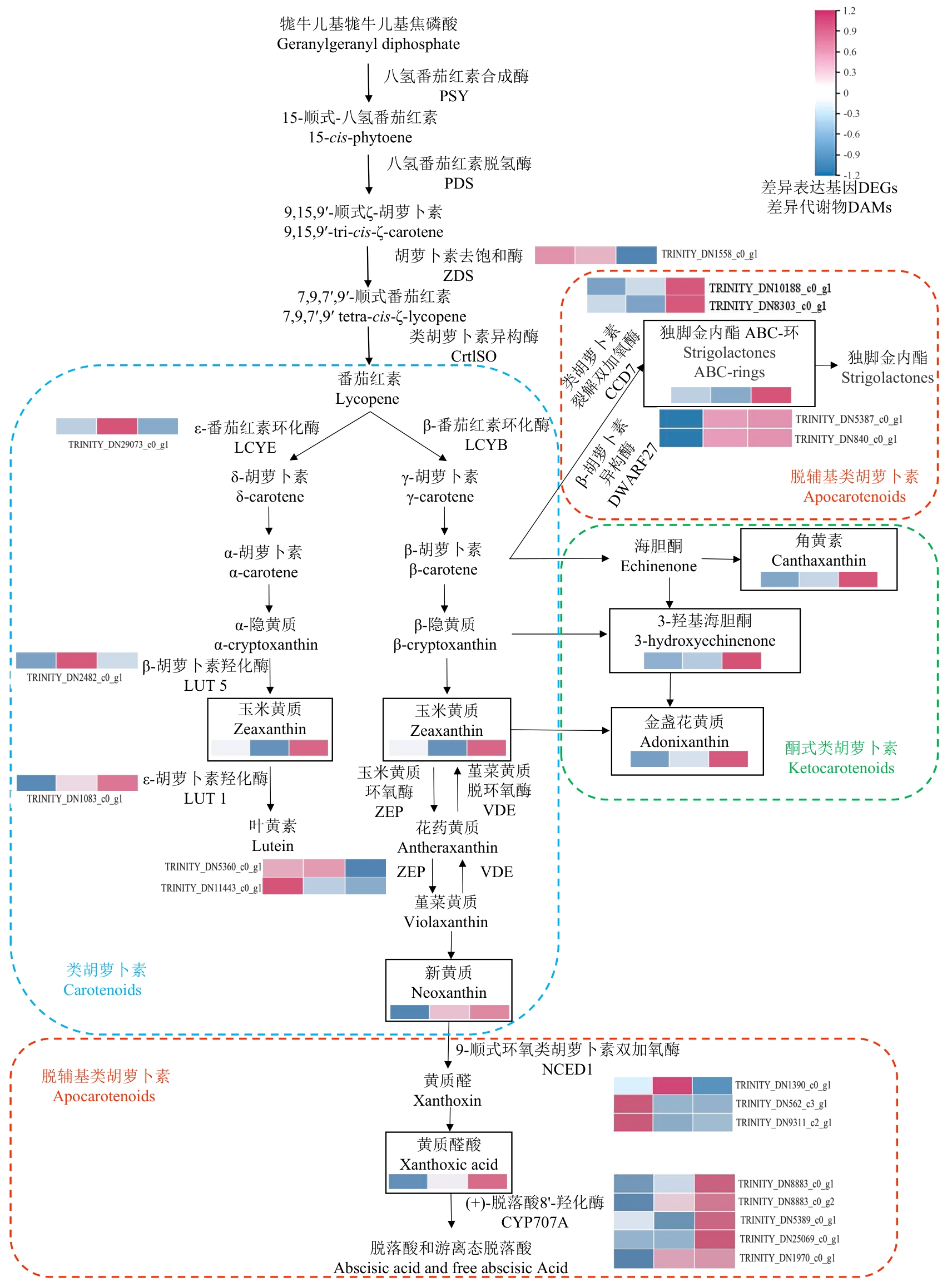
图4 不同遮光处理下金线莲叶片中类胡萝卜素的转录代谢联合分析热图的方块从左到右代表T1、T2、T3。不同颜色虚线框表示不同类型的类胡萝卜素
Fig. 4 Conjoint analysis of transcriptional metabolism of carotenoid in A. roxburghii leaves under different shading treatmentsIn the heatmap, the blocks from left to right correspond to the experimental treatment groups T1, T2, and T3. Different colored dashed boxes indicate different types of carotenoids

图5 不同遮光条件下金线莲类胡萝卜素相关转录因子与差异表达基因的调控关系圆形代表差异表达基因,方形代表转录因子
Fig. 5 Regulatory relationships between carotenoid-related transcription factors and differentially expressed genes in A. roxburghii under different shading conditionsCircles indicate differentially expressed genes, and squares indicate transcription factors

图7 八个差异表达基因的RT-qPCR分析不同的字母表示在0.05水平上差异显著(P<0.05)
Fig. 7 RT-qPCR analysis of eight differentially expressed genesDifferent letters indicate significant differences at the 0.05 level (P<0.05)
| 1 | 吕欣锴, 周丽思, 郭顺星. 我国金线兰资源特征及繁育技术研究进展 [J]. 药学学报, 2022, 57(7): 2057-2067. |
| Lü XK, Zhou LS, Guo SX. Resource characteristics and propagation techniques of Anoectochilus roxburghii in China [J]. Acta Pharm Sin, 2022, 57(7): 2057-2067. | |
| 2 | 王睿. 海南热带条件下金线莲种植适应性及规模化栽培试验研究 [D]. 海口: 海南大学, 2019. |
| Wang R. Experimental study on adaptability and large-scale cultivation of Anoectochilus roxburghii under tropical conditions in Hainan [D]. Haikou: Hainan University, 2019. | |
| 3 | Srivastava R. Physicochemical, antioxidant properties of carotenoids and its optoelectronic and interaction studies with chlorophyll pigments [J]. Sci Rep, 2021, 11(1): 18365. |
| 4 | Milani A, Basirnejad M, Shahbazi S, et al. Carotenoids: biochemistry, pharmacology and treatment [J]. Br J Pharmacol, 2017, 174(11): 1290-1324. |
| 5 | Bai C, Berman J, Farre G, et al. Reconstruction of the astaxanthin biosynthesis pathway in rice endosperm reveals a metabolic bottleneck at the level of endogenous β-carotene hydroxylase activity [J]. Transgenic Res, 2017, 26(1): 13-23. |
| 6 | 何静娟, 范燕萍. 观赏植物花色相关的类胡萝卜素组成及代谢调控研究进展 [J]. 园艺学报, 2022, 49(5): 1162-1172. |
| He JJ, Fan YP. Progress in composition and metabolic regulation of carotenoids related to floral color [J]. Acta Hortic Sin, 2022, 49(5): 1162-1172. | |
| 7 | Sathasivam R, Radhakrishnan R, Kim JK, et al. An update on biosynthesis and regulation of carotenoids in plants [J]. S Afr N J Bot, 2021, 140: 290-302. |
| 8 | Wang HT, Tian YC, Li YX, et al. Analysis of carotenoids and gene expression in apple germplasm resources reveals the role of MdCRTISO and MdLCYE in the accumulation of carotenoids [J]. J Agric Food Chem, 2023, 71(41): 15121-15131. |
| 9 | Li C, Wang CL, Cheng ZY, et al. Carotenoid biosynthesis genes LcLCYB, LcLCYE, and LcBCH from wolfberry confer increased carotenoid content and improved salt tolerance in tobacco [J]. Sci Rep, 2024, 14(1): 10586. |
| 10 | Du YX, Peng L, Dong B, et al. Genome-wide identification of 9-cis-epoxy-carotenoid dioxygenases (NCEDs) and potential function of OfNCED4 in carotenoid biosynthesis of Osmanthus fragrans [J]. Trees, 2024, 38(4): 891-902. |
| 11 | Sun TH, Rao S, Zhou XS, et al. Plant carotenoids: recent advances and future perspectives [J]. Mol Hortic, 2022, 2(1): 3. |
| 12 | 代绿叶, 张鑫, 滕英姿, 等. LED光照强度对紫叶生菜幼苗的影响 [J]. 中国农学通报, 2023, 39(10): 24-30. |
| Dai LY, Zhang X, Teng YZ, et al. Effects of LED light intensity on purple leaf lettuce seedlings [J]. Chin Agric Sci Bull, 2023, 39(10): 24-30. | |
| 13 | Formisano L, Ciriello M, El-Nakhel C, et al. Pearl grey shading net boosts the accumulation of total carotenoids and phenolic compounds that accentuate the antioxidant activity of processing tomato [J]. Antioxidants, 2021, 10(12): 1999. |
| 14 | Fu XM, Chen JM, Li JL, et al. Mechanism underlying the carotenoid accumulation in shaded tea leaves [J]. Food Chem X, 2022, 14: 100323. |
| 15 | 梁敏华, 梁瑞进, 杨震峰, 等. NAC6转录因子在芒果果实采后类胡萝卜素代谢过程中的表达及影响 [J]. 食品科学, 2024, 45(16): 77-87. |
| Liang MH, Liang RJ, Yang ZF, et al. Expression of NAC6 transcription factor and its impact on carotenoid metabolism in postharvest mango fruit [J]. Food Sci, 2024, 45(16): 77-87. | |
| 16 | Hu LP, Yang C, Zhang LN, et al. Effect of light-emitting diodes and ultraviolet irradiation on the soluble sugar, organic acid, and carotenoid content of postharvest sweet oranges (Citrus sinensis (L.) osbeck) [J]. Molecules, 2019, 24(19): 3440. |
| 17 | Xu YN, You CJ, Xu CB, et al. Red and blue light promote tomato fruit coloration through modulation of hormone homeostasis and pigment accumulation [J]. Postharvest Biol Technol, 2024, 207: 112588. |
| 18 | Jia DJ, Li YC, Jia K, et al. Abscisic acid activates transcription factor module MdABI5-MdMYBS1 during carotenoid-derived apple fruit coloration [J]. Plant Physiol, 2024, 195(3): 2053-2072. |
| 19 | Huang JF, Qin YL, Xie ZL, et al. Combined transcriptome and metabolome analysis reveal that the white and yellow mango pulp colors are associated with carotenoid and flavonoid accumulation, and phytohormone signaling [J]. Genomics, 2023, 115(5): 110675. |
| 20 | 魏翠华, 谢宇, 秦建彬, 等. 光照强度对金线莲生长及产量的影响 [J]. 北方园艺, 2015(12): 139-141. |
| Wei CH, Xie Y, Qin JB, et al. Effect of light intensity on the growth and yield of Anoectochilus roxburghii [J]. North Hortic, 2015(12): 139-141. | |
| 21 | 牛欢, 韦坤华, 徐倩, 等. 不同光照度对金线莲生长、生理特性和药用成分的影响 [J]. 植物资源与环境学报, 2020, 29(1): 26-36, 43. |
| Niu H, Wei KH, Xu Q, et al. Effects of different illuminances on growth, physiological characteristics, and medicinal components of Anoectochilus roxburghii [J]. J Plant Resour Environ, 2020, 29(1): 26-36, 43. | |
| 22 | Cao JY, Zeng JJ, Hu RQ, et al. Comparative metabolome and transcriptome analyses of the regulatory mechanism of light intensity in the synthesis of endogenous hormones and anthocyanins in Anoectochilus roxburghii (Wall.) Lindl [J]. Genes, 2024, 15(8): 989. |
| 23 | 陈常理, 李文略, 安霞, 等. 光照强度对金线莲多糖和黄酮类物质含量的影响 [J]. 分子植物育种, 2023, 21(15): 5103-5109. |
| Chen CL, Li WL, An X, et al. Effects of light intensities on polysaccharide and flavonoid contents in Anoectothenia roxburghii [J]. Mol Plant Breed, 2023, 21(15): 5103-5109. | |
| 24 | 程雅倩. 金线莲调节糖脂代谢活性及机制研究 [D]. 杭州: 浙江农林大学, 2024. |
| Cheng YQ. Study on the regulation activity and mechanism of Anoectochilus roxburghii on glycolipid metabolism [D]. Hangzhou: Zhejiang A & F University, 2024. | |
| 25 | 王立, 余翠婷. 金线莲和铁皮石斛无糖组培与传统组培的对照试验 [J]. 基因组学与应用生物学, 2020, 39(9): 4162-4170. |
| Wang L, Yu CT. A comparative experiment on sugar-free tissue culture and traditional tissue culture of Anemone roxburghii and Dendrobium candidum [J]. Genom Appl Biol, 2020, 39(9): 4162-4170. | |
| 26 | 林蔚, 王晶晶, 何官榕, 等. 金线莲不同栽培模式及不同组织的多糖含量 [J]. 福建农林大学学报: 自然科学版, 2020, 49(1): 40-44. |
| Lin W, Wang JJ, He GR, et al. Polysaccharide contents in different tissues of Anoectochilus roxburghii grown under various cultivation modes [J]. J Fujian Agric For Univ Nat Sci Ed, 2020, 49(1): 40-44. | |
| 27 | 钱承文, 罗栩辉, 林协全, 等. 腐殖质土添加杉木屑对金线莲生长及活性成分的影响 [J]. 中国野生植物资源, 2024, 43(3): 39-43. |
| Qian CW, Luo XH, Lin XQ, et al. Effect of Chinese fir sawdust added to humus soil on growth and main activities of Anoectochilus roxburghii [J]. Chin Wild Plant Resour, 2024, 43(3): 39-43. | |
| 28 | 赵琪, 王弛, 赵峰. 水培与泥炭土培金线莲生长及代谢差异比较 [J]. 亚热带植物科学, 2024, 53(1): 22-33. |
| Zhao Q, Wang C, Zhao F. Differences in growth and metabolics between hydroponic and peat soil cultivation Anoectochilus roxburghii [J]. Subtrop Plant Sci, 2024, 53(1): 22-33. | |
| 29 | Sonobe R, Miura Y, Sano T, et al. Estimating leaf carotenoid contents of shade-grown tea using hyperspectral indices and PROSPECT-D inversion [J]. Int J Remote Sens, 2018, 39(5): 1306-1320. |
| 30 | 高新征, 黄东爱, 邬强, 等. 虾青素生产及其生物合成途径的研究进展 [J]. 海南医学院学报, 2013, 19(1): 141-144. |
| Gao XZ, Huang DA, Wu Q, et al. Research progress on astaxanthin production and its biosynthesis pathway [J]. J Hainan Med Univ, 2013, 19(1): 141-144. | |
| 31 | 梁婉凤, 曾菁菁, 胡若群, 等. 转录组与代谢组分析不同生长时期金线莲类胡萝卜素的积累 [J]. 生物技术通报, 2024, 40(10): 262-274. |
| Liang WF, Zeng JJ, Hu RQ, et al. Transcriptional and metabolomic analysis of carotenoid accumulation in Anoectochilus roxburghii during different growth periods [J]. Biotechnol Bull, 2024, 40(10): 262-274. | |
| 32 | Fu XM, Cheng SH, Feng C, et al. Lycopene cyclases determine high α-/β-carotene ratio and increased carotenoids in bananas ripening at high temperatures [J]. Food Chem, 2019, 283: 131-140. |
| 33 | Richaud D, Stange C, Gadaleta A, et al. Identification of Lycopene Epsilon cyclase (LCYE) gene mutants to potentially increase β-carotene content in durum wheat (Triticum turgidum L.ssp. durum) through TILLING [J]. PLoS One, 2018, 13(12): e0208948. |
| 34 | Kim J, DellaPenna D. Defining the primary route for lutein synthesis in plants: the role of Arabidopsis carotenoid beta-ring hydroxylase CYP97A3 [J]. Proc Natl Acad Sci U S A, 2006, 103(9): 3474-3479. |
| 35 | Schwarz N, Armbruster U, Iven T, et al. Tissue-specific accumulation and regulation of Zeaxanthin epoxidase in Arabidopsis reflect the multiple functions of the enzyme in plastids [J]. Plant Cell Physiol, 2015, 56(2): 346-357. |
| 36 | Lee SY, Jang SJ, Jeong HB, et al. A mutation in Zeaxanthin epoxidase contributes to orange coloration and alters carotenoid contents in pepper fruit (Capsicum annuum) [J]. Plant J, 2021, 106(6): 1692-1707. |
| 37 | Ye SH, Huang YY, Ma TT, et al. BnaABF3 and BnaMYB44 regulate the transcription of zeaxanthin epoxidase genes in carotenoid and abscisic acid biosynthesis [J]. Plant Physiol, 2024, 195(3): 2372-2388. |
| 38 | Ahrazem O, Rubio-Moraga A, Berman J, et al. The carotenoid cleavage dioxygenase CCD2 catalysing the synthesis of crocetin in spring crocuses and saffron is a plastidial enzyme [J]. New Phytol, 2016, 209(2): 650-663. |
| 39 | 周煌, 张议斤, 罗志雄, 等. 水稻NCED基因家族生物信息学分析 [J]. 分子植物育种, 2022, 20(4): 1060-1067. |
| Zhou H, Zhang YJ, Luo ZX, et al. Bioinformatics analysis of NCED gene family in rice [J]. Mol Plant Breed, 2022, 20(4): 1060-1067. | |
| 40 | 张泽华, 万淑媛, 李琴, 等. 桃PpNCED家族成员鉴定与表达特性分析 [J]. 西北农业学报, 2021, 30(10): 1495-1503. |
| Zhang ZH, Wan SY, Li Q, et al. Identification and expression analysis of PpNCED family membersin Prunus persica [J]. Acta Agric Boreali Occidentalis Sin, 2021, 30(10): 1495-1503. | |
| 41 | Zhu F, Luo T, Liu CY, et al. An R2R3-MYB transcription factor represses the transformation of α- and β-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulate [J]. New Phytol, 2017, 216(1): 178-192. |
| 42 | Song HY, Liu JH, Chen CQ, et al. Down-regulation of NCED leads to the accumulation of carotenoids in the flesh of F1 generation of peach hybrid [J]. Front Plant Sci, 2022, 13: 1055779. |
| 43 | Wen K, Li XL, Yin T, et al. Genome-wide identification of carotenoid cleavage oxygenase genes in Orah mandarin and the mechanism by which CrCCD4b1 affects peel color [J]. Sci Hortic, 2024, 338: 113652. |
| 44 | Abuauf H, Haider I, Jia KP, et al. The Arabidopsis DWARF27 gene encodes an all-trans-/ 9-cis-β-carotene isomerase and is induced by auxin, abscisic acid and phosphate deficiency [J]. Plant Sci, 2018, 277: 33-42. |
| 45 | Bruno M, Al-Babili S. On the substrate specificity of the rice strigolactone biosynthesis enzyme DWARF27 [J]. Planta, 2016, 243(6): 1429-1440. |
| 46 | 肖玉洁, 李泽明, 易鹏飞, 等. 转录因子参与植物低温胁迫响应调控机理的研究进展 [J]. 生物技术通报, 2018, 34(12): 1-9. |
| Xiao YJ, Li ZM, Yi PF, et al. Research progress on response mechanism of transcription factors involved in plant cold stress [J]. Biotechnol Bull, 2018, 34(12): 1-9. | |
| 47 | Liu F, Xi MW, Liu T, et al. The central role of transcription factors in bridging biotic and abiotic stress responses for plants' resilience [J]. New Crops, 2024, 1: 100005. |
| 48 | 周俊杰, 王艺光, 董彬, 等. 桂花OfPSY、OfPDS和OfHYB基因启动子克隆及表达特性分析 [J]. 浙江农林大学学报, 2023, 40(1): 64-71. |
| Zhou JJ, Wang YG, Dong B, et al. Cloning and expression characterization of OfPSY, OfPDS and OfHYB gene promoters in Osmanthus fragrans [J]. J Zhejiang A F Univ, 2023, 40(1): 64-71. | |
| 49 | Sun Q, He ZC, Wei RR, et al. The transcriptional regulatory module CsHB5-CsbZIP44 positively regulates abscisic acid-mediated carotenoid biosynthesis in Citrus (Citrus spp.) [J]. Plant Biotechnol J, 2024, 22(3): 722-737. |
| 50 | Sun Q, He ZC, Feng D, et al. The abscisic acid-responsive transcriptional regulatory module CsERF110-CsERF53 orchestrates Citrus fruit coloration [J]. Plant Commun, 2024, 5(11): 101065. |
| 51 | Wu C, Sun L, Lv YZ, et al. Functional characterization and in silico analysis of phytoene synthase family genes responsible for carotenoid biosynthesis in watermelon (Citrullus lanatus L.) [J]. Agronomy, 2020, 10(8): 1077. |
| [1] | 刘源, 赵冉, 卢振芳, 李瑞丽. 植物类胡萝卜素生物代谢途径及其功能研究进展[J]. 生物技术通报, 2025, 41(5): 23-31. |
| [2] | 罗嗣芳, 张祖铭, 谢丽芳, 郭紫晶, 陈兆星, 杨月华, 严翔, 张洪铭. 山金柑GATA基因家族全基因组鉴定及在果实发育中的表达分析[J]. 生物技术通报, 2025, 41(5): 218-230. |
| [3] | 杨朝结, 张兰, 陈红, 黄娟, 石桃雄, 朱丽伟, 陈庆富, 李洪有, 邓娇. 苦荞转录因子基因FtbHLH3调控类黄酮生物合成的功能鉴定[J]. 生物技术通报, 2025, 41(4): 134-144. |
| [4] | 王天禧, 杨炳松, 潘荣君, 盖文贤, 梁美霞. 苹果PLATZ基因家族鉴定及MdPLATZ9基因功能研究[J]. 生物技术通报, 2025, 41(4): 176-187. |
| [5] | 宋姝熠, 蒋开秀, 刘欢艳, 黄亚成, 刘林娅. ‘红阳’猕猴桃TCP基因家族鉴定及其在果实中的表达分析[J]. 生物技术通报, 2025, 41(3): 190-201. |
| [6] | 李旭娟, 李纯佳, 刘洪博, 徐超华, 林秀琴, 陆鑫, 刘新龙. 甘蔗腋芽形成发育过程的转录组分析[J]. 生物技术通报, 2025, 41(3): 202-218. |
| [7] | 王斌, 王玉昆, 肖艳辉. 丁香罗勒(Ocimum gratissimum)叶片响应镉胁迫的比较转录组学分析[J]. 生物技术通报, 2025, 41(3): 255-270. |
| [8] | 刘洁, 王飞, 陶婷, 张玉静, 陈浩婷, 张瑞星, 石玉, 张毅. 过表达SlWRKY41提高番茄幼苗抗旱性[J]. 生物技术通报, 2025, 41(2): 107-118. |
| [9] | 赵长延, 柳延涛, 贾秀苹, 刘胜利, 雷中华, 王鹏, 朱志锋, 董红业, 吕增帅, 段维, 万素梅. 盐碱胁迫下褪黑素对作物生理机制影响的研究进展[J]. 生物技术通报, 2025, 41(2): 18-29. |
| [10] | 李艳伟, 杨妍妍, 孙亚玲, 霍雨猛, 王振宝, 刘冰江. 基于转录组分析植物激素对洋葱鳞茎膨大发育的调控机制[J]. 生物技术通报, 2025, 41(2): 187-201. |
| [11] | 钱政毅, 吴绍芳, 曹舒怡, 宋雅欣, 潘鑫峰, 李兆伟, 范凯. 睡莲NAC转录因子的鉴定及其表达分析[J]. 生物技术通报, 2025, 41(2): 234-247. |
| [12] | 寇焙森, 程萌萌, 郭雪琴, 葛彬, 刘迪, 陆海, 李慧. 组蛋白去乙酰化酶抑制剂TSA处理对杨树茎生长发育的影响[J]. 生物技术通报, 2025, 41(1): 240-251. |
| [13] | 裴旭娟, 狄靖宜, 刘浩, 高伟霞. 基于转录组分析挖掘兽疫链球菌透明质酸分子量调控元件[J]. 生物技术通报, 2025, 41(1): 347-356. |
| [14] | 沈川, 李夏, 覃剑锋, 段龙飞, 刘佳. 基于软腐病菌诱导的魔芋酵母双杂交文库筛选WRKY72互作蛋白[J]. 生物技术通报, 2025, 41(1): 85-94. |
| [15] | 马博涛, 伍国强, 魏明. bZIP转录因子在植物逆境胁迫响应和生长发育中的作用[J]. 生物技术通报, 2024, 40(9): 148-160. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||