








生物技术通报 ›› 2026, Vol. 42 ›› Issue (1): 241-250.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0482
收稿日期:2025-05-13
出版日期:2026-01-26
发布日期:2025-12-23
通讯作者:
晁跃辉,男,教授,研究方向 :草类植物遗传育种;E-mail: chaoyuehui@bjfu.edu.cn作者简介:张驰昊,男,硕士研究生,研究方向 :草类植物遗传育种;E-mail: zzzch0127@126.com基金资助:
ZHANG Chi-hao( ), LIU Jin-nan(
), LIU Jin-nan( ), CHAO Yue-hui(
), CHAO Yue-hui( )
)
Received:2025-05-13
Published:2026-01-26
Online:2025-12-23
摘要:
目的 克隆蒺藜苜蓿(Medicago truncatula)MtbZIP29基因,研究bZIP29转录因子转录自激活、亚细胞定位及表达特征等,为后续阐明MtbZIP29基因参与蒺藜苜蓿生长发育、内源激素信号转导提供一定的理论研究基础。 方法 从野生型苜蓿‘R108’中克隆MtbZIP29基因,构建试验所需的表达载体,通过瞬时表达融合蛋白观察其亚细胞定位,利用酵母表达分析其自激活活性;对MtbZIP29基因进行生物信息学分析,包括蛋白质理化性质、启动子顺式作用元件、蛋白质结构等;利用RT-qPCR方法对不同组织中及不同激素(ABA、SA、6-BA、IAA、MeJA)处理条件下的MtbZIP29进行表达分析;利用农杆菌介导法,获得转基因烟草植株,分析MtbZIP29的功能。 结果 成功克隆MtbZIP29基因,基因编码区全长1 518 bp,编码506个氨基酸,分子量55.830 kD,理论等电点为6.82,不稳定系数63.60,为不稳定亲水性蛋白;二级结构预测α螺旋占比25.89%,延伸链为0.59%,无规则卷曲为73.52%;亚细胞定位结果表明,该蛋白定位于细胞核;酵母实验表明,该基因编码蛋白具有转录自激活活性;表达特征分析显示,MtbZIP29基因在叶片中的表达水平最高,而茎和荚果中的表达水平明显低于其他组织,同时MtbZIP29基因的表达水平明显受到不同激素处理的影响。对转基因和野生型植株对比发现,转基因烟草的根系更加发达,同时根茎处具有明显的膨大。 结论 MtbZIP29基因参与根系生长发育,对不同激素均有响应,可能通过整合激素信号参与植物根系形态建成及逆境适应性的调控过程。
张驰昊, 刘晋囡, 晁跃辉. 蒺藜苜蓿bZIP转录因子MtbZIP29的克隆及功能分析[J]. 生物技术通报, 2026, 42(1): 241-250.
ZHANG Chi-hao, LIU Jin-nan, CHAO Yue-hui. Cloning and Functional Analysis of a bZIP Transcription Factor MtbZIP29 from Medicago truncatula[J]. Biotechnology Bulletin, 2026, 42(1): 241-250.
引物名称 Primer name | 序列 Sequence(5′-3′) | 用途 Usage |
|---|---|---|
| MtbZIP29-F | GATAAGCGCCTGCTCCTT | 基因克隆 Gene cloning |
| MtbZIP29-R | ACGACCTCCAATCCAACC | |
| M13-47 | CAGGGTTTTCCCAGTCACG | |
| M13-48 | GAGCGGATAACAATTTCACAC | |
| 3302-bZIP29-F | CATGGTAGATCTGACTAGTATGGAAGATACTGAAGCAGCTTCATCTTTCAAAATG | 构建亚细胞定位载体 Construction of subcellular localization vector |
| 3302-bZIP29-R | GGCCTCCTAGGTACACGCGTTTGCTTTAACTCATTCTTTGAGTTGGCATT | |
| BDbZIP29-F | TCAGAGGAGGACCTGCATATGGAAGATACTGAAGCAGCT | 构建酵母表达载体 Construction of yeast expression vector |
| BD-bZIP29-R | CGCTGCAGGTCGACGGATCCCTATTGCTTTAACTCATTCTTTGAGTTG | |
| BD-bZIP29-U-F | GCCATGGAGGCCGAATTCCCGATGGAAGATACTGAAGCAGC | 构建酵母表达载体(N端) Construction of yeast expression vector (N-terminus) |
| BD-ZIP29-U-R | CTGCAGGTCGACGGATCCCCTTACACAACTTCACCTTCCGGCG | |
| BD-bZIP29-D-F | GCCATGGAGGCCGAATTCCCGGATGATCTTTTTTCTGCTTA | 构建酵母表达载体(C端) Construction of yeast expression vector (C-terminus) |
| BD-bZIP29-D-R | CTGCAGGTCGACGGATCCCCCTATTGCTTTAACTCATTCT | |
| MtbZIP29 qRT-F | TGGGAATGGTGAGTTTAG | 基因表达分析 Gene expression analysis |
| MtbZIP29 qRT-R | CAGAAAGTGTAGTGGCTTC | |
| MtActin-F | TGATCTGGCTGGTCGTGACCTTA | 蒺藜苜蓿内参基因表达分析 Expression analysis of Medicago truncatula internal reference gene |
| MtActin-R | ATGCCTGCTGCTTCCATTCCTAT | |
| Nt-bZIP29-F | ATCAGATCCCGGTTTCAC | 转基因烟草PCR检测 PCR analysis of transgenic tobacco |
| Nt-bZIP29-R | CTATCATCAGTAGCCGCGT |
表1 引物及序列
Table 1 Primers and sequences
引物名称 Primer name | 序列 Sequence(5′-3′) | 用途 Usage |
|---|---|---|
| MtbZIP29-F | GATAAGCGCCTGCTCCTT | 基因克隆 Gene cloning |
| MtbZIP29-R | ACGACCTCCAATCCAACC | |
| M13-47 | CAGGGTTTTCCCAGTCACG | |
| M13-48 | GAGCGGATAACAATTTCACAC | |
| 3302-bZIP29-F | CATGGTAGATCTGACTAGTATGGAAGATACTGAAGCAGCTTCATCTTTCAAAATG | 构建亚细胞定位载体 Construction of subcellular localization vector |
| 3302-bZIP29-R | GGCCTCCTAGGTACACGCGTTTGCTTTAACTCATTCTTTGAGTTGGCATT | |
| BDbZIP29-F | TCAGAGGAGGACCTGCATATGGAAGATACTGAAGCAGCT | 构建酵母表达载体 Construction of yeast expression vector |
| BD-bZIP29-R | CGCTGCAGGTCGACGGATCCCTATTGCTTTAACTCATTCTTTGAGTTG | |
| BD-bZIP29-U-F | GCCATGGAGGCCGAATTCCCGATGGAAGATACTGAAGCAGC | 构建酵母表达载体(N端) Construction of yeast expression vector (N-terminus) |
| BD-ZIP29-U-R | CTGCAGGTCGACGGATCCCCTTACACAACTTCACCTTCCGGCG | |
| BD-bZIP29-D-F | GCCATGGAGGCCGAATTCCCGGATGATCTTTTTTCTGCTTA | 构建酵母表达载体(C端) Construction of yeast expression vector (C-terminus) |
| BD-bZIP29-D-R | CTGCAGGTCGACGGATCCCCCTATTGCTTTAACTCATTCT | |
| MtbZIP29 qRT-F | TGGGAATGGTGAGTTTAG | 基因表达分析 Gene expression analysis |
| MtbZIP29 qRT-R | CAGAAAGTGTAGTGGCTTC | |
| MtActin-F | TGATCTGGCTGGTCGTGACCTTA | 蒺藜苜蓿内参基因表达分析 Expression analysis of Medicago truncatula internal reference gene |
| MtActin-R | ATGCCTGCTGCTTCCATTCCTAT | |
| Nt-bZIP29-F | ATCAGATCCCGGTTTCAC | 转基因烟草PCR检测 PCR analysis of transgenic tobacco |
| Nt-bZIP29-R | CTATCATCAGTAGCCGCGT |
| 用途 Usage | 网址 Website |
|---|---|
| 启动子及顺式作用元件预测 Promoter and cis-acting element prediction | PlantCARE, http://bioinformatics.psb.ugent.be/webtools/plantcare/html |
| 系统进化树分析 Phylogenetic tree analysis | NCBI, https://www.ncbi.nlm.nih.gov/ |
| 蛋白质结构域分析 Protein structure domain analysis | UniProt, https://www.uniprot.org/ |
| 蛋白质二级结构预测 Protein secondary structure prediction | SOPMA, https://npsa.lyon.inserm.fr/cgibin/npsa_automat.pl page=/NPSA/npsa_sopma.html |
| 蛋白质三级结构预测 Protein tertiary structure prediction | Swissmodel, https://swissmodel.expasy.org/ |
| 蛋白质理化性质分析 Analysis of physicochemical properties of proteins | Expasy-ProtParam, http://web.expasy.org/protparam Expasy-ProtScale, http://web.expasy.org/protscale |
表2 MtbZIP29生物信息学分析
Table 2 Bioinformation analysis of MtbZIP29
| 用途 Usage | 网址 Website |
|---|---|
| 启动子及顺式作用元件预测 Promoter and cis-acting element prediction | PlantCARE, http://bioinformatics.psb.ugent.be/webtools/plantcare/html |
| 系统进化树分析 Phylogenetic tree analysis | NCBI, https://www.ncbi.nlm.nih.gov/ |
| 蛋白质结构域分析 Protein structure domain analysis | UniProt, https://www.uniprot.org/ |
| 蛋白质二级结构预测 Protein secondary structure prediction | SOPMA, https://npsa.lyon.inserm.fr/cgibin/npsa_automat.pl page=/NPSA/npsa_sopma.html |
| 蛋白质三级结构预测 Protein tertiary structure prediction | Swissmodel, https://swissmodel.expasy.org/ |
| 蛋白质理化性质分析 Analysis of physicochemical properties of proteins | Expasy-ProtParam, http://web.expasy.org/protparam Expasy-ProtScale, http://web.expasy.org/protscale |
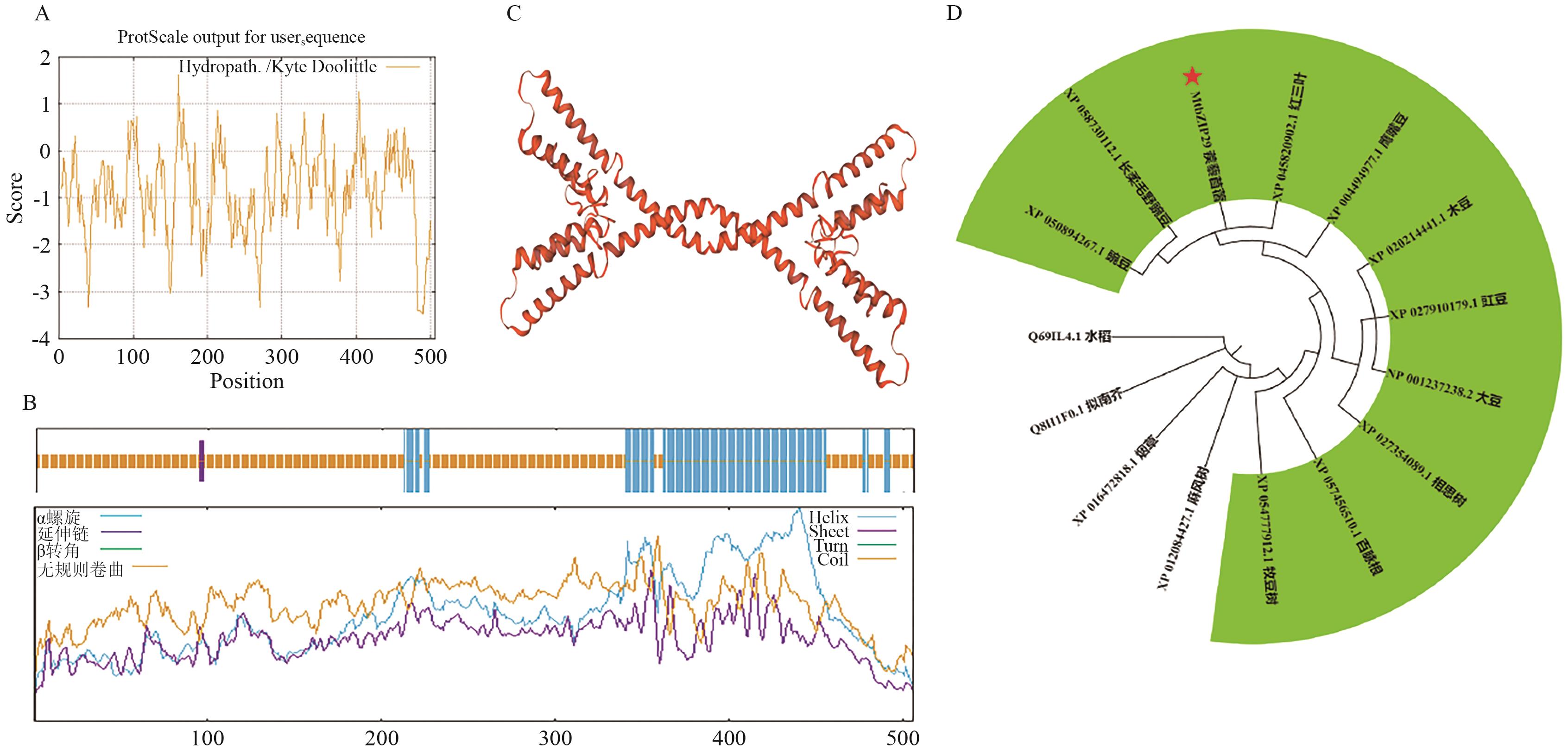
图3 MtbZIP29蛋白结构及遗传进化分析A:蛋白质亲疏水性分析;B:蛋白质二级结构;C:蛋白质三级结构;D:系统进化树分析
Fig. 3 Protein structure and genetic evolutionary analysis of protein MtbZIP29A: Hydrophobicity analysis of protein. B: The secondary structure of protein. C: The tertiary structure of protein. D: Phylogenetic tree analysis
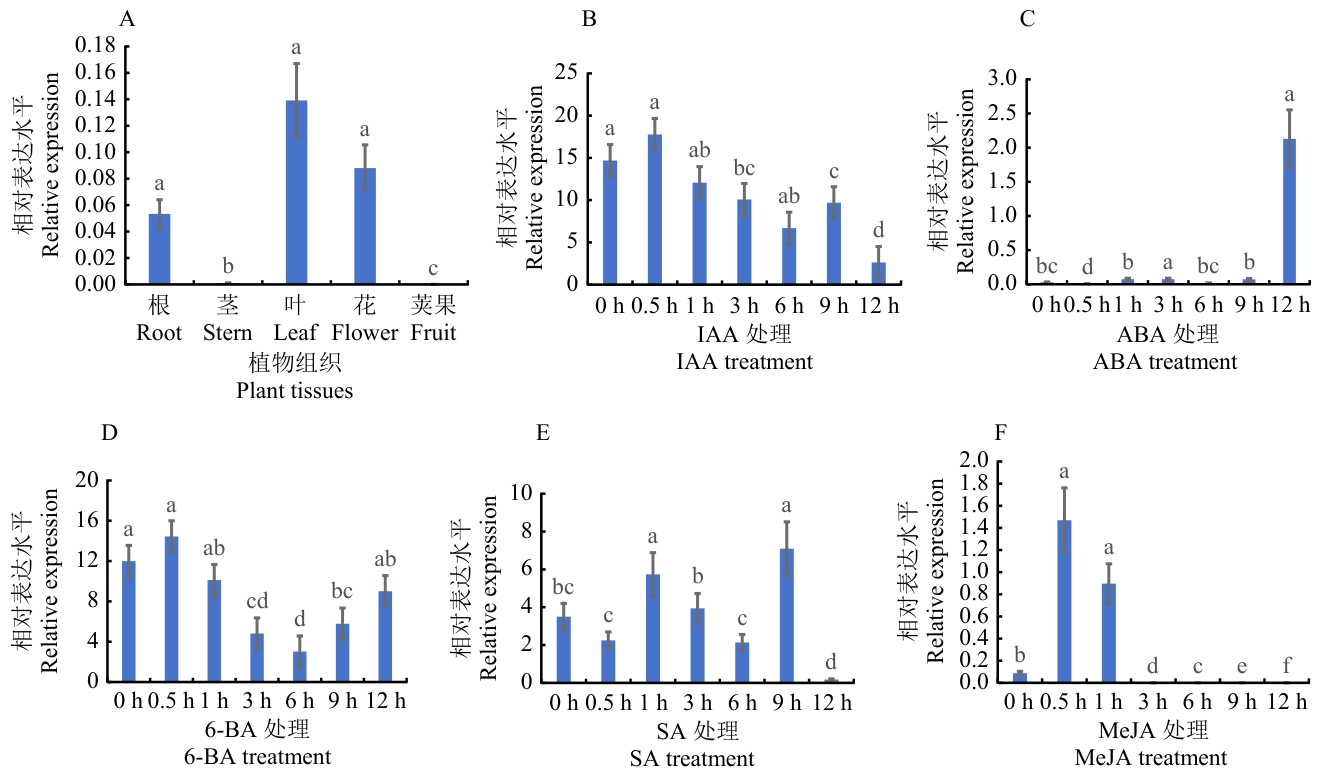
图4 MtbZIP表达分析不同字母代表P < 0.05差异显著,数值为平均值±标准误(n = 3)
Fig. 4 Expression analysis of MtbZIPDifferent letters indicate significant differences at P < 0.05. The values are in means ± SE (n = 3)
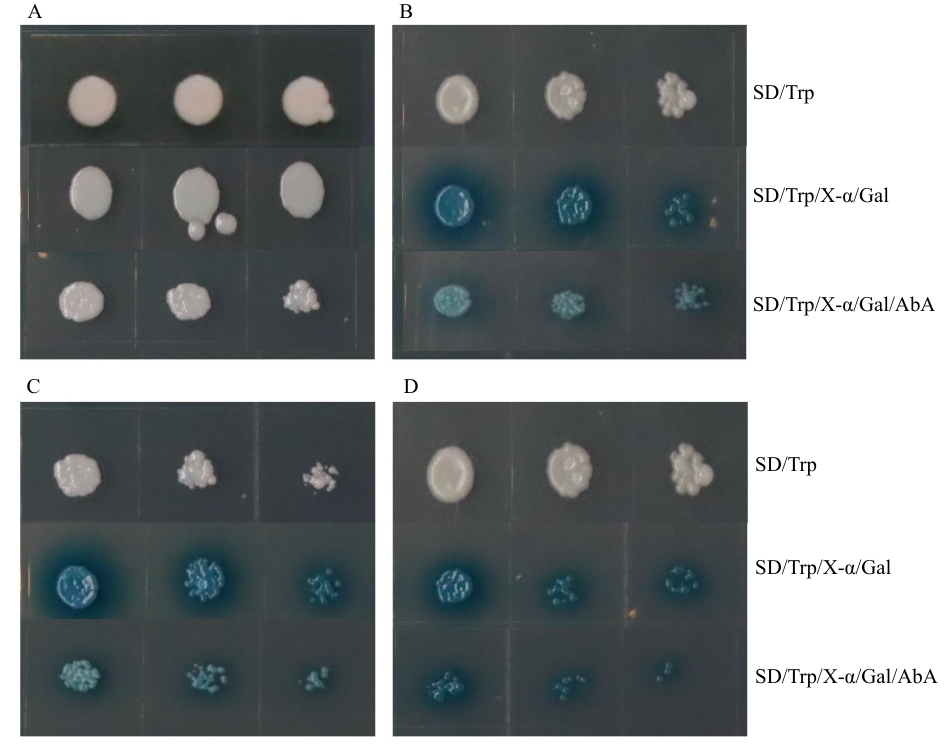
图6 MtbZIP29 自激活检测A: BD空载转化酵母菌;B:BD-bZIP29转化酵母菌;C: BD-bZIP29-N转化酵母菌;D: BD-bZIP29-C转化酵母菌
Fig. 6 MtbZIP29 self-activation assayA: Yeast transformed with BD empty vector. B: Yeast transformed with BD-bZIP29. C: Yeast transformed with BD-bZIP29-N. D: Yeast transformed with BD-bZIP29-C
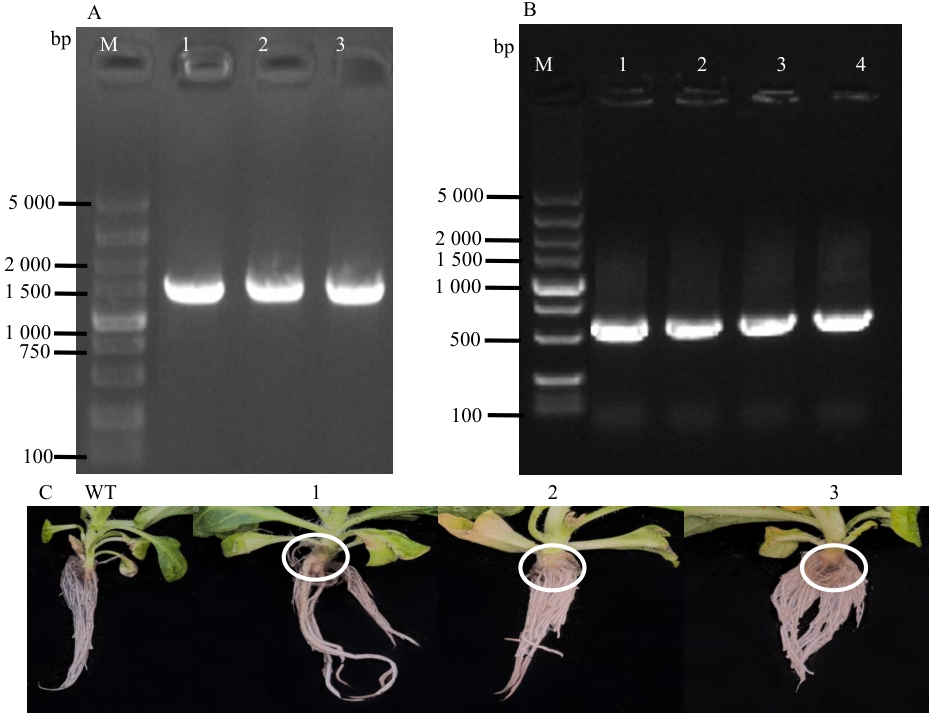
图8 GV3101转化、转基因烟草PCR及转基因植株观察A:3302-bZIP29转化GV3101(M:DL5000 DNA marker,1-3:GV3101菌落);B:转基因烟草PCR(M:DL5000 DNA marker,1-4:转基因烟草);C:转基因烟草表型图(WT为空白对照,1-3为转基因植株)
Fig. 8 Observation of GV3101 transformation, transgenic tobacco PCR, and transgenic plantA: 3302-bZIP29 transformed GV3101(M: DL5,000 DNA marker, 1-3: GV3101 colony). B: Transgenic tobacco PCR (M: DL5,000 DNA marker, 1-4: transgenic tobacco). C: Phenotype of transgenic tobacco (WT: wild type, 1-3: transgenic tobaccos)
| [1] | Weidemüller P, Kholmatov M, Petsalaki E, et al. Transcription factors: Bridge between cell signaling and gene regulation [J]. Proteomics, 2021, 21(23/24): 2000034. |
| [2] | Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins [J]. Science, 1988, 240(4860): 1759-1764. |
| [3] | Zhang TT, Zhou Y. Plant transcription factors and salt stress [M]//Plant Transcription Factors. Academic Press, 2023: 369-381. |
| [4] | Kuravsky M, Grey G, Kelly C F, et al. A basic leucine zipper uses a dimer pathway to locate its targets in DNA mixtures[J]. bioRxiv, 2025: 2025.08. 25.672131. |
| [5] | Yu Y, Qian YC, Jiang MY, et al. Regulation mechanisms of plant basic leucine zippers to various abiotic stresses [J]. Front Plant Sci, 2020, 11: 1258. |
| [6] | Wang BX, Xu B, Liu Y, et al. A Novel mechanisms of the signaling cascade associated with the SAPK10-bZIP20-NHX1 synergistic interaction to enhance tolerance of plant to abiotic stress in rice (Oryza sativa L.) [J]. Plant Sci, 2022, 323: 111393. |
| [7] | Hussain Q, Asim M, Zhang R, et al. Transcription factors interact with ABA through gene expression and signaling pathways to mitigate drought and salinity stress [J]. Biomolecules, 2021, 11(8): 1159. |
| [8] | Yoshida T, Fujita Y, Sayama H, et al. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation [J]. Plant J, 2010, 61(4): 672-685. |
| [9] | Manna M, Thakur T, Chirom O, et al. Transcription factors as key molecular target to strengthen the drought stress tolerance in plants [J]. Physiol Plant, 2021, 172(2): 847-868. |
| [10] | 曹丽茹, 马晨晨, 庞芸芸, 等. 玉米bZIP转录因子ZmbZIP26的分子特征、亚细胞定位及响应非生物胁迫的表达分析 [J]. 华北农学报, 2023, 38(4): 1-10. |
| Cao LR, Ma CC, Pang YY, et al. Molecular characteristics, subcellular localization and expression analysis of maize bZIP transcription factor ZmbZIP26 in response to abiotic stress [J]. Acta Agric Boreali Sin, 2023, 38(4): 1-10. | |
| [11] | 郭纯, 宋桂梅, 闫艳, 等. 西洋参bZIP基因家族全基因组鉴定和表达分析 [J]. 生物技术通报, 2024, 40(4): 167-178. |
| Guo C, Song GM, Yan Y, et al. Genome wide identification and expression analysis of the bZIP gene family in Panax quinquefolius [J]. Biotechnol Bull, 2024, 40(4): 167-178. | |
| [12] | Gai WX, Ma X, Qiao YM, et al. Characterization of the bZIP transcription factor family in pepper (Capsicum annuum L.): CabZIP25 positively modulates the salt tolerance [J]. Front Plant Sci, 2020, 11: 139. |
| [13] | 张蕾, 陈昕涛, 白丽娟, 等. 大豆bZIP转录因子基因GmbZIP33的克隆与功能分析 [J]. 大豆科学, 2023, 42(5): 524-531. |
| Zhang L, Chen XT, Bai LJ, et al. Cloning and functional analysis of GmbZIP33 encoding bZIP transcription factor from soybean [J]. Soybean Sci, 2023, 42(5): 524-531. | |
| [14] | Li MY, Hwarari D, Li Y, et al. The bZIP transcription factors in Liriodendron chinense: Genome-wide recognition, characteristics and cold stress response [J]. Front Plant Sci, 2022, 13: 1035627. |
| [15] | Hsieh TH, Li CW, Su RC, et al. A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response [J]. Planta, 2010, 231(6): 1459-1473. |
| [16] | 曾力旺, 张小燕, 林墁, 等. 木薯MebZIP27基因的克隆与原核表达分析 [J]. 福建农业学报, 2025, 40(1): 1-9. |
| Zeng LW, Zhang XY, Lin M, et al. Cloning and prokaryotic expression analysis of cassava MebZIP27 gene [J]. Fujian J Agric Sci, 2025, 40(1): 1-9. | |
| [17] | 曹晓宇, 王学敏, 郭继承, 等. 紫花苜蓿bZIP转录因子MsbZIP1的克隆、表达特性和遗传转化分析 [J]. 草地学报, 2020, 28(3): 613-622. |
| Cao XY, Wang XM, Guo JC, et al. Cloning, expression characteristics and genetic transformation analysis of a bZIP transcription factor MsbZIP1 in alfalfa [J]. Acta Agrestia Sin, 2020, 28(3): 613-622. | |
| [18] | Liu SX, Qin B, Fang QX, et al. Genome-wide identification, phylogeny and expression analysis of the bZIP gene family in Alfalfa (Medicago sativa) [J]. Biotechnol Biotechnol Equip, 2021, 35(1): 905-916. |
| [19] | Zhao K, Chen S, Yao WJ, et al. Genome-wide analysis and expression profile of the bZIP gene family in poplar [J]. BMC Plant Biol, 2021, 21(1): 122. |
| [20] | Dash M, Yordanov YS, Georgieva T, et al. Poplar PtabZIP1-like enhances lateral root formation and biomass growth under drought stress [J]. Plant J, 2017, 89(4): 692-705. |
| [21] | 赵安琪, 尹跃, 何军, 等. 枸杞LbaHY5基因克隆、亚细胞定位及表达分析 [J]. 华北农学报, 2024, 39(6): 76-83. |
| Zhao AQ, Yin Y, He J, et al. Cloning, subcellular localization and expression analysis of LbaHY5 gene from Lycium barbarum [J]. Acta Agric Boreali Sin, 2024, 39(6): 76-83. | |
| [22] | Yang SS, Zhang XX, Zhang XM, et al. A bZIP transcription factor, PqbZIP1, is involved in the plant defense response of American ginseng [J]. PeerJ, 2022, 10: e12939. |
| [23] | Niu XL, Fu DQ. The roles of BLH transcription factors in plant development and environmental response [J]. Int J Mol Sci, 2022, 23(7): 3731. |
| [24] | Verma S, Attuluri VPS, Robert HS. Transcriptional control of Arabidopsis seed development [J]. Planta, 2022, 255(4): 90. |
| [25] | Guo ZL, Dzinyela R, Yang LM, et al. bZIP transcription factors: structure, modification, abiotic stress responses and application in plant improvement [J]. Plants, 2024, 13(15): 2058. |
| [26] | Alonso-Blanco C, Aarts MGM, Bentsink L, et al. What has natural variation taught us about plant development, physiology, and adaptation? [J]. Plant Cell, 2009, 21(7): 1877-1896. |
| [27] | Li JS, Wang XM, Zhang YL, et al. cGMP regulates hydrogen peroxide accumulation in calcium-dependent salt resistance pathway in Arabidopsis thaliana roots [J]. Planta, 2011, 234(4): 709-722. |
| [28] | Kaur P, Lui C, Dudchenko O, et al. Delineating the Tnt1 insertion landscape of the model legume Medicago truncatula cv. R108 at the hi-C resolution using a chromosome-length genome assembly [J]. Int J Mol Sci, 2021, 22(9): 4326. |
| [29] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method [J]. Methods, 2001, 25(4): 402-408. |
| [30] | Riechmann JL, Heard J, Martin G, et al. Arabidopsis Transcription factors: genome-wide comparative analysis among eukaryotes [J]. Science, 2000, 290(5499): 2105-2110. |
| [31] | Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis [J]. Plant Cell, 2007, 19(2): 485-494. |
| [32] | Yoshida T, Fujita Y, Maruyama K, et al. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress [J]. Plant Cell Environ, 2015, 38(1): 35-49. |
| [33] | Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress [J]. Proc Natl Acad Sci U S A, 2009, 106(20): 8380-8385. |
| [34] | Cutler SR, Rodriguez PL, Finkelstein RR, et al. Abscisic acid: emergence of a core signaling network [J]. Annu Rev Plant Biol, 2010, 61: 651-679. |
| [35] | Luang S, Sornaraj P, Bazanova N, et al. The wheat TabZIP2 transcription factor is activated by the nutrient starvation-responsive SnRK3/CIPK protein kinase [J]. Plant Mol Biol, 2018, 96(6): 543-561. |
| [36] | Han XY, Wei XP, Lu WJ, et al. Transcriptional regulation of KCS gene by bZIP29 and MYB70 transcription factors during ABA-stimulated wound suberization of kiwifruit (Actinidia deliciosa) [J]. BMC Plant Biol, 2022, 22(1): 23. |
| [37] | Zhang LN, Zhang LC, Xia C, et al. A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis [J]. Physiol Plant, 2015, 153(4): 538-554. |
| [38] | Van Leene J, Blomme J, Kulkarni SR, et al. Functional characterization of the Arabidopsis transcription factor bZIP29 reveals its role in leaf and root development [J]. J Exp Bot, 2016, 67(19): 5825-5840. |
| [1] | 程婷婷, 刘俊, 王利丽, 练从龙, 魏文君, 郭辉, 吴尧琳, 杨晶凡, 兰金旭, 陈随清. 杜仲查尔酮异构酶基因家族全基因组鉴定及其表达模式分析[J]. 生物技术通报, 2025, 41(9): 242-255. |
| [2] | 翟莹, 计俊杰, 陈炯辛, 于海伟, 李珊珊, 赵艳, 马天意. 异源过表达大豆GmNF-YB24提高转基因烟草抗旱性[J]. 生物技术通报, 2025, 41(8): 137-145. |
| [3] | 张泽, 杨秀丽, 宁东贤. 花生4CL基因家族鉴定及对干旱与盐胁迫响应分析[J]. 生物技术通报, 2025, 41(7): 117-127. |
| [4] | 黄旭升, 周雅莉, 柴旭东, 闻婧, 王计平, 贾小云, 李润植. 紫苏质体型PfLPAT1B基因的克隆及其在油脂合成中的功能分析[J]. 生物技术通报, 2025, 41(7): 226-236. |
| [5] | 董栩焜, 车永梅, 王明硕, 罗政刚, 管恩森, 赵方贵, 叶青, 刘新. 纳米硅和蜡样芽胞杆菌SS1共处理对烟草生长的影响[J]. 生物技术通报, 2025, 41(7): 292-298. |
| [6] | 杨春, 王晓倩, 王红军, 晁跃辉. 蒺藜苜蓿MtZHD4基因克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2025, 41(5): 244-254. |
| [7] | 卢勇杰, 夏海乾, 李永铃, 张文建, 余婧, 赵会纳, 王兵, 许本波, 雷波. 烟草AP2/ERF转录因子NtESR2的克隆及功能分析[J]. 生物技术通报, 2025, 41(4): 266-277. |
| [8] | 季盈彤, 高嘉宁, 钱孟英, 潘宁, 张筠, 崔红, 闫筱筱. NtMYC2调控烟草表皮毛发育的功能研究[J]. 生物技术通报, 2025, 41(4): 278-288. |
| [9] | 赵春夺, 李玉娥, 刘友杰, 王新航, 赵伟, 黄永成, 李虎林, 姬文秀. 轮作与连作对烟草根际土壤养分、酶活性及微生物群落结构的影响[J]. 生物技术通报, 2025, 41(4): 312-322. |
| [10] | 马耀武, 张麒宇, 杨淼, 蒋诚, 张振宇, 张伊琳, 李梦莎, 许嘉阳, 张斌, 崔光周, 姜瑛. 烟草根际促生菌的筛选鉴定及促生性能研究[J]. 生物技术通报, 2025, 41(3): 271-281. |
| [11] | 韩梦荞, 吴疆, 李丽华, 王昭懿, 邓习, 韦凤杰, 任民, 孙洋洋, 李富欣. 烟草染色体制备体系的建立与优化[J]. 生物技术通报, 2025, 41(3): 44-50. |
| [12] | 蒋天威, 李亚娇, 马培杰, 陈才俊, 刘晓霞, 陈莹, 王小利. 蒺藜苜蓿开花过程中的全基因组DNA甲基化分析[J]. 生物技术通报, 2025, 41(11): 301-310. |
| [13] | 冯小虎, 张文梅, 徐婧, 熊书斌, 王利兵, 宋文静. 一组靶向烟草青枯病的噬菌体鸡尾酒筛选与防控效果[J]. 生物技术通报, 2025, 41(11): 311-318. |
| [14] | 王碧成, 景海青, 万坤, 张莹莹, 丁家豪, 李润植, 薛金爱, 张海平. 大豆BCAT基因家族鉴定及GmBCAT3在干旱胁迫中的功能分析[J]. 生物技术通报, 2025, 41(10): 196-209. |
| [15] | 李粘前, 李琛, 李淑婷, 马菊花, 景海青, 孙岩, 周雅莉, 薛金爱, 李润植. 油莎豆苹果酸酶(ME)全基因组鉴定及CeNAD-ME2功能分析[J]. 生物技术通报, 2025, 41(10): 210-221. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||