








生物技术通报 ›› 2025, Vol. 41 ›› Issue (12): 240-253.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0534
杨涛1,2( ), 李琳1, 莫小连1, 陈晓龙1, 王健1, 黄园1, 赵杰宏1, 邹颉1(
), 李琳1, 莫小连1, 陈晓龙1, 王健1, 黄园1, 赵杰宏1, 邹颉1( )
)
收稿日期:2025-05-24
出版日期:2025-12-26
发布日期:2026-01-06
通讯作者:
邹颉,男,博士,副教授,研究方向 :中药民族药资源生产与开发利用;E-mail: zxgh202@whu.edu.cn作者简介:杨涛,男,硕士研究生,研究方向 :中药民族药资源生产与开发利用;E-mail: 1586613641@qq.com
基金资助:
YANG Tao1,2( ), LI Lin1, MO Xiao-lian1, CHEN Xiao-long1, WANG Jian1, HUANG Yuan1, ZHAO Jie-hong1, ZOU Jie1(
), LI Lin1, MO Xiao-lian1, CHEN Xiao-long1, WANG Jian1, HUANG Yuan1, ZHAO Jie-hong1, ZOU Jie1( )
)
Received:2025-05-24
Published:2025-12-26
Online:2026-01-06
摘要:
目的 鉴定铁皮石斛DELLA基因家族主效基因DoDELLA2在植物形态发育中的功能,并研究DoDELLA2基因调控的下游基因及信号通路,为铁皮石斛分子育种提供理论依据。 方法 采用RT-qPCR对铁皮石斛DELLA基因家族成员进行组织表达量分析,筛选到铁皮石斛DELLA基因家族潜在主效基因DoDELLA2,通过全基因合成DoDELLA2并构建35S-DoDELLA2过表达载体异源转化拟南芥,结合表型观察与转录组数据分析开展功能研究。 结果 过表达DoDELLA2拟南芥与野生型相比,抽薹时间延迟10-16 d;株高显著降低40.04%;叶片叶尖呈椭圆形且边缘锯齿加深;角果数量显著增多但长度显著缩短等生育期表型差异。转录组分析表明,过表达DoDELLA2拟南芥与野生型相比,共有696个差异基因,其中上调基因中差异显著且富集的主要有转运体蛋白、赤霉素合成酶、富含半胱氨酸分泌蛋白等蛋白编码基因。下调基因中差异显著且富集的主要有过氧化物酶蛋白超家族、细胞色素P450蛋白超家族、植物病程相关蛋白(PR1)等。差异基因显著富集于木质素生物合成、营养生长向生殖生长转变等生物学过程,以及苯丙烷生物合成、类黄酮生物合成、二萜生物合成等KEGG通路,且赤霉素生物合成关键酶基因AtGA20OX2、AtGA20OX3和AtGA3OX1表达上调表现出明显的反馈调控。 结论 过表达DoDELLA2基因通过调控GA信号通路下游基因影响拟南芥的形态发育,DoDELLA2基因过表达显著影响赤霉素合成酶基因和部分抗性基因家族表达,为铁皮石斛品质与性状改良的分子育种实践提供重要靶点和理论基础。
杨涛, 李琳, 莫小连, 陈晓龙, 王健, 黄园, 赵杰宏, 邹颉. 铁皮石斛DoDELLA2的功能研究[J]. 生物技术通报, 2025, 41(12): 240-253.
YANG Tao, LI Lin, MO Xiao-lian, CHEN Xiao-long, WANG Jian, HUANG Yuan, ZHAO Jie-hong, ZOU Jie. Functional Study of DoDELLA2 in Dendrobium officinale Kimura et Migo[J]. Biotechnology Bulletin, 2025, 41(12): 240-253.
| 引物名称 Primer name | 上游引物 Forward primer (5′-3′) | 下游引物 Reverse primer (5′-3′) |
|---|---|---|
| DoDELLA2 | ACTAGGGTCTCGCACCATGAAGAGGGAGCATTTGGAGAGTGTTGGAGGAA | ACTAGGGTCTCTCGCCGTGAGCATCTGCGGCCGCGGAAGCGGA |
| 35S | CACGGGGGACTCTTGCCACC | GACACGCTGAACTTGTGG |
| eGFP | — | GACACGCTGAACTTGTGG |
| DoDELLA2-CDS | ATGAAGAGGGAGCATTTGGAG | GTGAGCATCTGCGGCCGCGGA |
| DoDELLA2-RT | CTCCTGTTGTCTTCCCTGATTT | TCGGGTTCCACTATCGATCT |
| AtActin | TCAGATGCCCAGAAGTCTTGTTCC | CCGTACAGATCCTTCCTGATATCC |
| DoDELLA1-RT | CAAGAGAGTGGAAGCGGATTAG | ATCGGCATAGCTGACGAATAAA |
| DoDELLA3-RT | CGGAGTCCTTGCATTACTACTC | CGAGAAATACCTCCGACATCAC |
| DoDELLA4-RT | CGAGGCACTGCACTTCTATT | CCGAGATAGACTTCCGACATAAAC |
| DoActin | TCCCAAGGCAAACAGAGAAA | GGCCACTAGCATATAGGGAAAG |
| AtGA2OX7 | AAACCTTGCTTGGAAGCCCT | TCACGCTTTGGTACACTCC |
| AtGASA1 | TCTCCAACTCGTCCAGGCTGATG | CTACACACGCACTCCCACAATCG |
| AtGALS2 | ACGACGGGATAGGAAGTATGCGG | TCCCCTTCCGCTCTGTGATACGTC |
| AtGA20OX2 | GCAGAAGCTTGCACCAAACA | GTGGAGAATCTGCCGGTGAA |
| AtGA20OX3 | ATCAAGACCAAGTTGGCGGT | TAGAGCCATGAAGGTGTCGC |
| AtGA3OX1 | GATCTCCTCTTCTCCGCTGC | TTTGGAAGGCACCCCAAGTT |
表1 引物序列
Table 1 Primer sequence
| 引物名称 Primer name | 上游引物 Forward primer (5′-3′) | 下游引物 Reverse primer (5′-3′) |
|---|---|---|
| DoDELLA2 | ACTAGGGTCTCGCACCATGAAGAGGGAGCATTTGGAGAGTGTTGGAGGAA | ACTAGGGTCTCTCGCCGTGAGCATCTGCGGCCGCGGAAGCGGA |
| 35S | CACGGGGGACTCTTGCCACC | GACACGCTGAACTTGTGG |
| eGFP | — | GACACGCTGAACTTGTGG |
| DoDELLA2-CDS | ATGAAGAGGGAGCATTTGGAG | GTGAGCATCTGCGGCCGCGGA |
| DoDELLA2-RT | CTCCTGTTGTCTTCCCTGATTT | TCGGGTTCCACTATCGATCT |
| AtActin | TCAGATGCCCAGAAGTCTTGTTCC | CCGTACAGATCCTTCCTGATATCC |
| DoDELLA1-RT | CAAGAGAGTGGAAGCGGATTAG | ATCGGCATAGCTGACGAATAAA |
| DoDELLA3-RT | CGGAGTCCTTGCATTACTACTC | CGAGAAATACCTCCGACATCAC |
| DoDELLA4-RT | CGAGGCACTGCACTTCTATT | CCGAGATAGACTTCCGACATAAAC |
| DoActin | TCCCAAGGCAAACAGAGAAA | GGCCACTAGCATATAGGGAAAG |
| AtGA2OX7 | AAACCTTGCTTGGAAGCCCT | TCACGCTTTGGTACACTCC |
| AtGASA1 | TCTCCAACTCGTCCAGGCTGATG | CTACACACGCACTCCCACAATCG |
| AtGALS2 | ACGACGGGATAGGAAGTATGCGG | TCCCCTTCCGCTCTGTGATACGTC |
| AtGA20OX2 | GCAGAAGCTTGCACCAAACA | GTGGAGAATCTGCCGGTGAA |
| AtGA20OX3 | ATCAAGACCAAGTTGGCGGT | TAGAGCCATGAAGGTGTCGC |
| AtGA3OX1 | GATCTCCTCTTCTCCGCTGC | TTTGGAAGGCACCCCAAGTT |
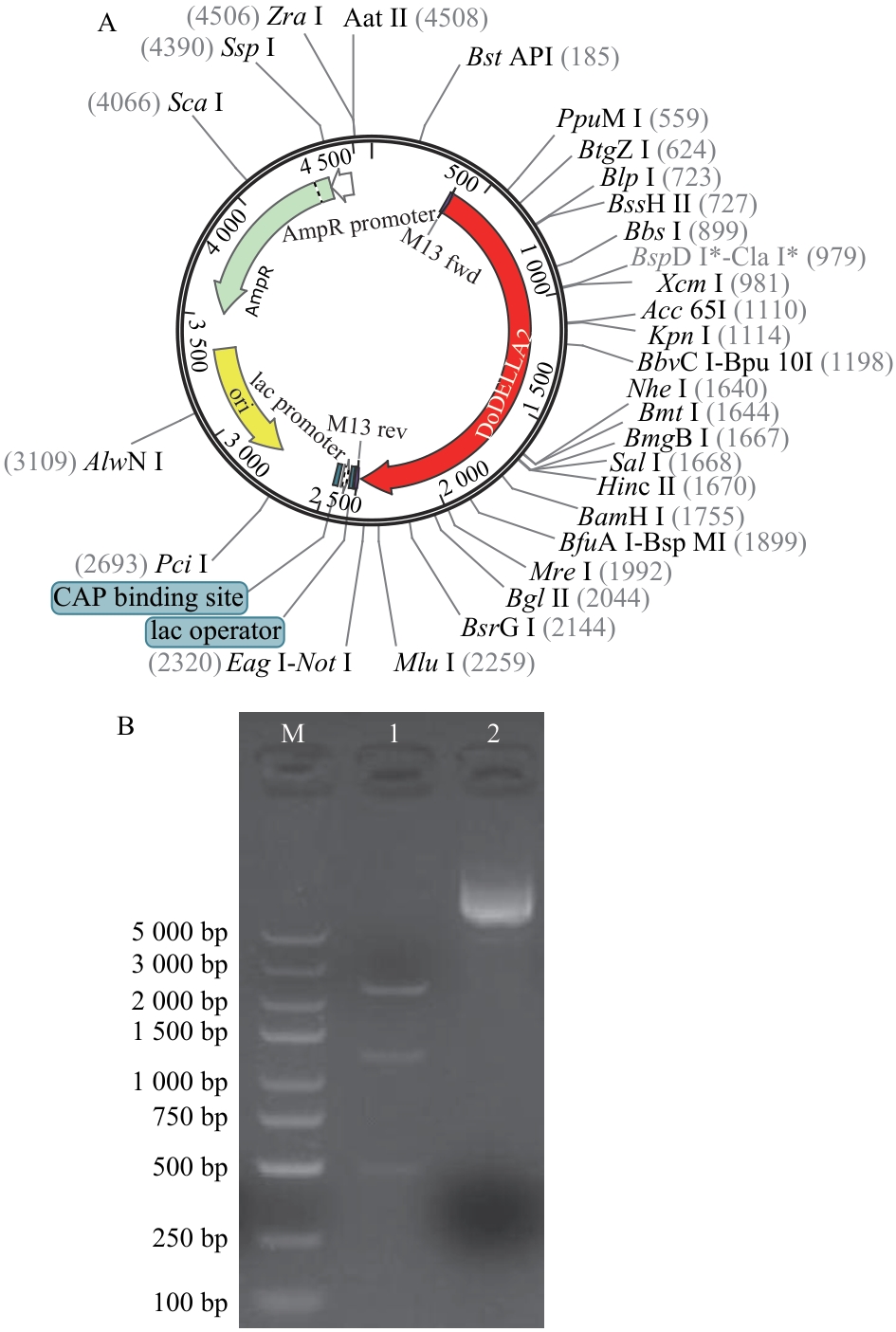
图3 pUC57-DoDELLA2质粒酶切验证A:pUC57-DoDELLA2质粒图谱;B:质粒酶切验证(1:ApaL I酶切pUC57-DoDELLA2质粒;2:pUC57-DoDELLA2质粒;M:DL 5 000 maker)
Fig. 3 Enzyme digestion verification of pUC57-DoDELLA2 plasmidA: Plasmid map of pUC57-DoDELLA2. B: Plasmid enzyme digestion verification (1: pUC57-DoDELLA2 plasmid digested with ApaL I; 2: pUC57-DoDELLA2 plasmid; M: DL 5 000 marker)
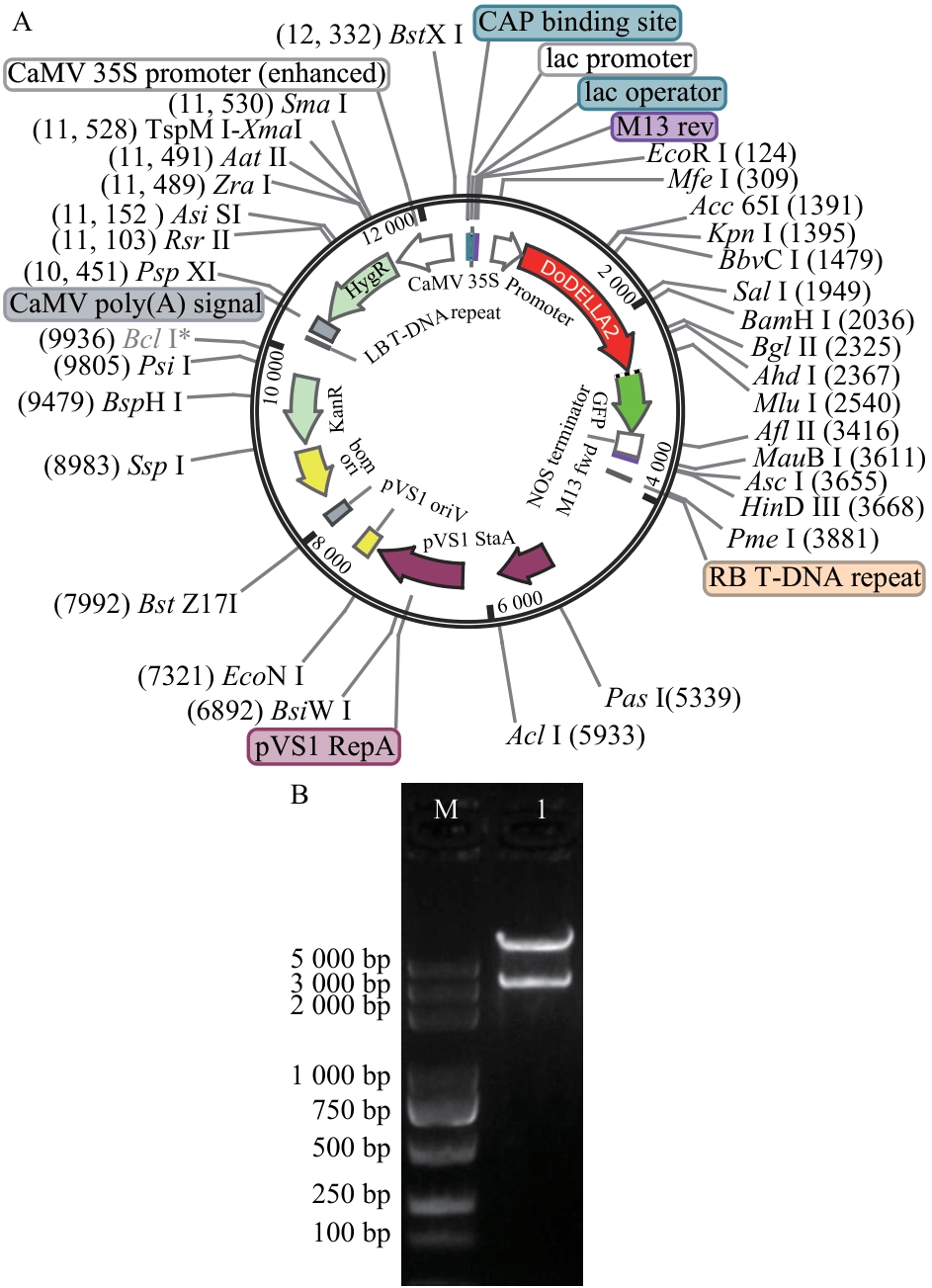
图4 pEGOEP35S-H-DoDELLA2-GFP质粒酶切验证A:pEGOEP35S-H-DoDELLA2-GFP质粒图谱;B:EcoR I/Hind III对质粒pEGOEP35S-H-DoDELLA2-GFP进行酶切验证;M:DL 5 000 maker
Fig. 4 Enzyme digestion verification of pEGOEP35S-H-DoDELLA2-GFP plasmidA: Plasmid map of pEGOEP35S-H-DoDELLA2-GFP. B: Restriction enzyme digestion verification of plasmid pEGOEP35S-H-DoDELLA2-GFP with EcoR I/Hind III. M: DL 5 000 marker
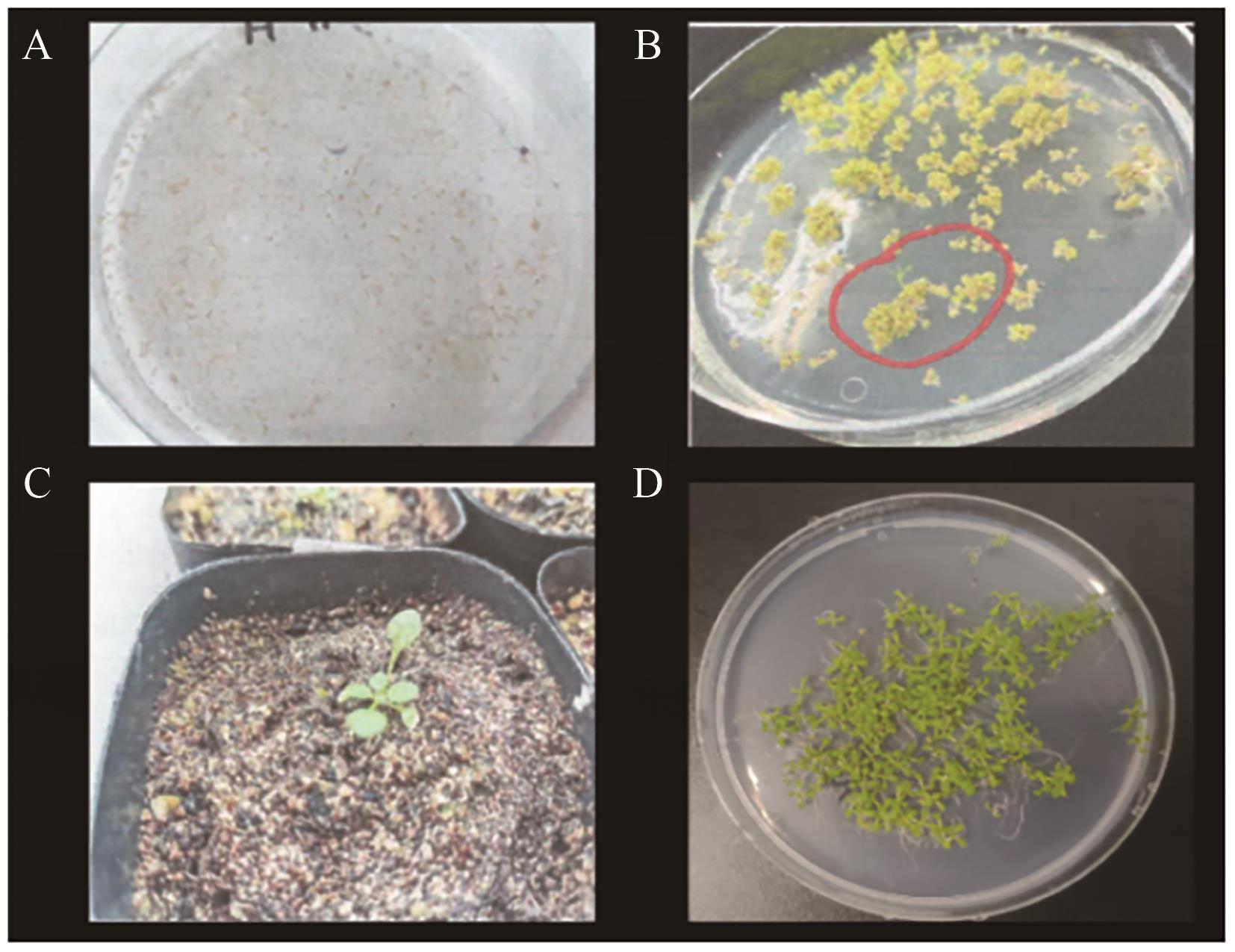
图5 过表达DoDELLA2拟南芥阳性苗的筛选A:T0代的种子;B:阳性苗的筛选;C:阳性苗移栽;D:分离比的筛选
Fig. 5 Screening of positive seedlings for A. thaliana overexpressing DoDELLA2A: Seeds of the T0 generation. B: Screening of positive seedlings. C: Transplantation of positive seedlings. D: Screening of segregation ratio
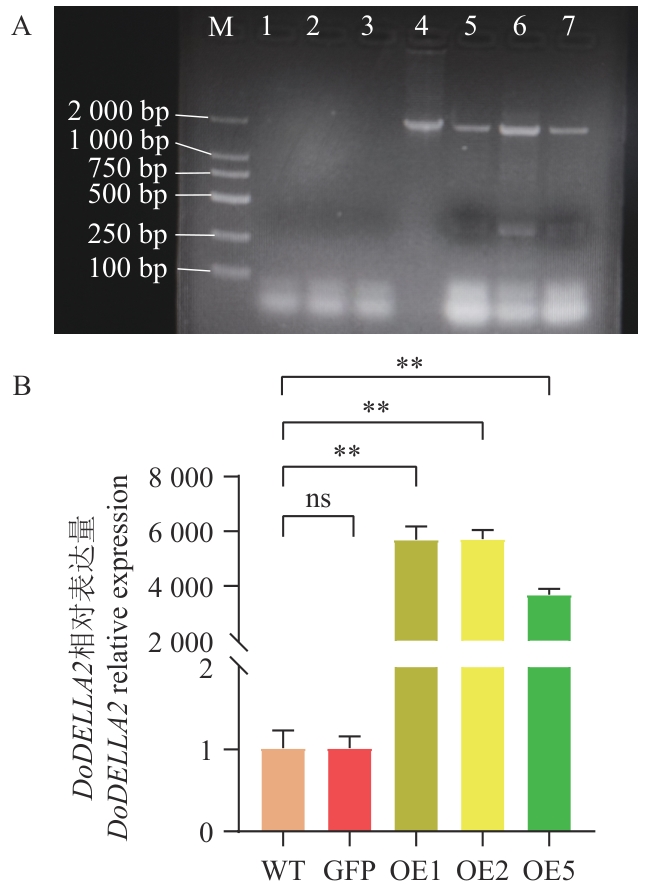
图6 过表达DoDELLA2拟南芥的PCR鉴定及DoDELLA2的表达情况A:过表达DoDELLA2拟南芥的cDNA PCR电泳图(1:水;2:WT;3:空载GFP拟南芥;4:阳性质粒;5‒7:分别为OE1、OE2和OE5;M:DL 2 000 maker);B:过表达DoDELLA2拟南芥的DoDELLA2相对表达量(WT:野生型拟南芥;GFP:空载拟南芥;OE1/2/5:过表达DoDELLA2拟南芥纯合株系);**P<0.01;下同
Fig. 6 PCR identification of A. thaliana overexpressing DoDELLA2 and expression analysis of DoDELLA2A: cDNA PCR electrophoresis profile of A. thaliana overexpressing DoDELLA2. (1: Water; 2: WT; 3: A. thaliana with empty GFP vector; 4: positive plasmid; 5‒7: OE1, OE2 and OE5, respectively; M: DL 2 000 marker). B: Relative expression of DoDELLA2 in A. thaliana overexpressing DoDELLA2 (WT: Wild-type A. thaliana; GFP: A. thaliana with empty vector; OE1/2/5: homozygous lines of A. thaliana overexpressing DoDELLA2 respectively). **P<0.01. The same below

图7 过表达DoDELLA2拟南芥抽薹及叶片表型观察A:8周龄拟南芥正面;B:8周龄拟南芥叶片;C:8周龄拟南芥俯视图;D:拟南芥抽薹天数;n=12
Fig. 7 Bolting and leaf phenotypic observation of A. thaliana overexpressing DoDELLA2A: Front view of 8-week-old A. thaliana. B: Leaves of 8-week-old A. thaliana. C: Top view of 8-week-old A. thaliana. D: Bolting days of A. thaliana. n=12

图8 过表达DoDELLA2拟南芥株高及角果表型观察A:12周龄拟南芥;B:9周龄拟南芥角果;C:12周龄拟南芥角果;D:12周龄拟南芥株高;E:12周龄拟南芥角果数量;n=12;*P<0.05
Fig. 8 Plant height and silique phenotypic observation of A. thaliana overexpressing DoDELLA2A: 12-week-old A. thaliana. B: Siliques of 9-week-old A. thaliana. C: Siliques of 12-week-old A. thaliana. D: Plant height of 12-week-old A. thaliana. E: Number of siliques of 12-week-old A. thaliana. n=12. *P<0.05
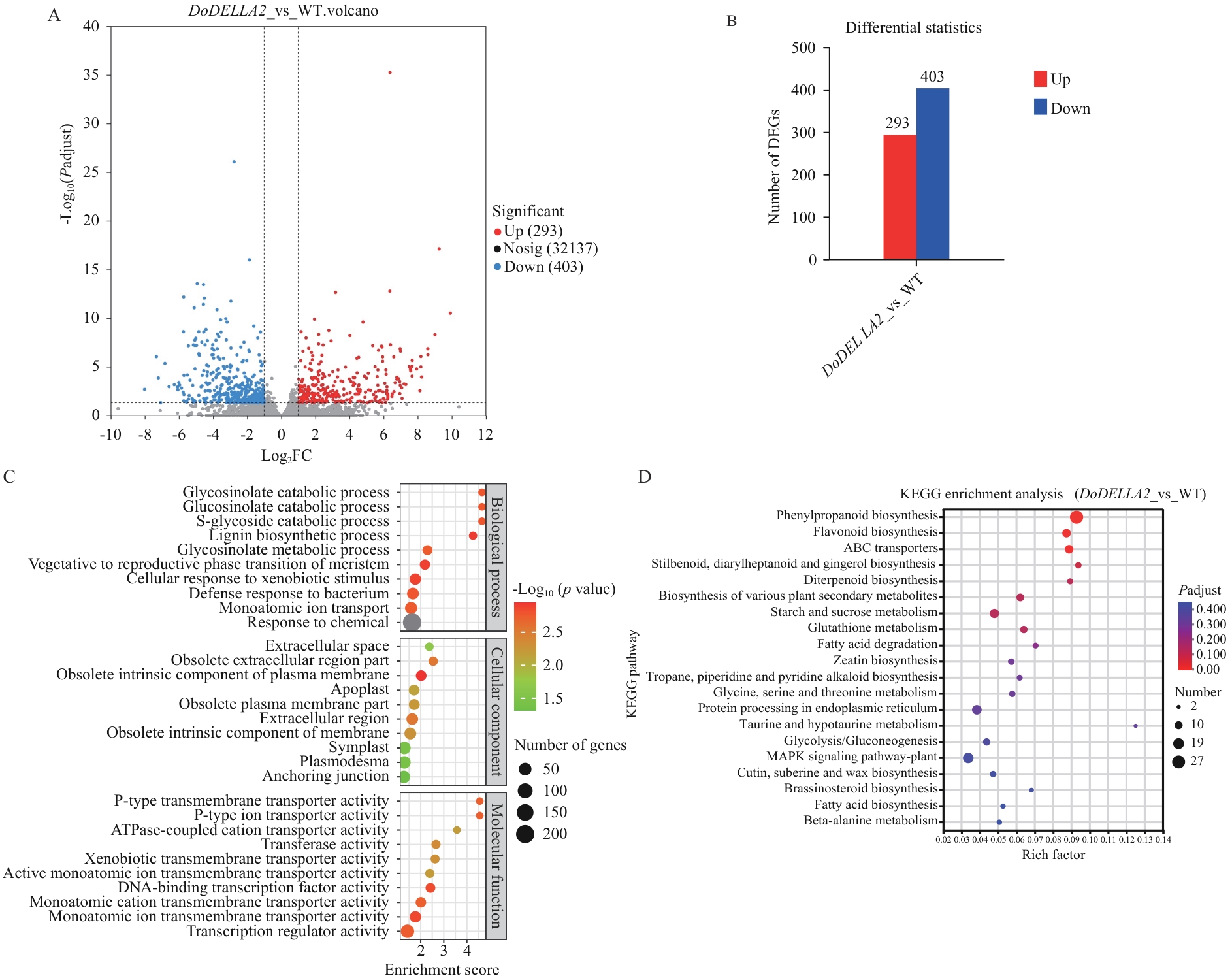
图9 铁皮石斛DoDELLA2异源转化拟南芥差异基因数据分析A:DoDELLA2_vs_WT火山图;B:DoDELLA2_vs_WT差异基因统计柱状图;C:DoDELLA2_vs_WT差异基因GO注释;D:DoDELLA2_vs_WT差异基因KEGG富集分析
Fig. 9 Differential gene data analysis of heterologous transformation of D. officinaleDoDELLA2 into A. thalianaA: Volcano plot of DoDELLA2_vs_WT. B: Statistical histogram of differentially expressed genes in DoDELLA2_vs_WT. C: GO annotation of differentially expressed genes in DoDELLA2_vs_WT. D: KEGG enrichment analysis of differentially expressed genes in DoDELLA2_vs_WT
基因名 Gene name | 基因描述 Gene description | 调控 Regulate |
|---|---|---|
| AT1G71680 | Transmembrane amino acid transporter family protein | Up |
| STP6 | Sugar transporter 6 | Up |
| PMT2 | Polyol/monosaccharide transporter 2 | Up |
| ACA7 | Cation transporter/E1-E2 ATPase family protein | Up |
| STP9 | Sugar transporter 9 | Up |
| CAT7 | Cationic amino acid transporter 7 | Up |
| AST91 | Sulfate transporter 91 | Up |
| PHT3;2 | Phosphate transporter 3;2 | Up |
| LHT1 | Lysine histidine transporter 1 | Up |
| ABCG1 | ABC-2 type transporter family protein | Up |
| GA20OX3 | Gibberellin 20-oxidase 3 | Up |
| GA20OX2 | Gibberellin 20 oxidase 2 | Up |
| GA3OX1 | Gibberellin 3-oxidase 1 | Up |
| AT3G21970 | Cysteine-rich repeat secretory protein, putative (DUF26) | Up |
| AT4G20530 | Cysteine-rich repeat secretory-like protein | Up |
| AT4G20530 | Cysteine-rich repeat secretory-like protein | Up |
| AT4G20530 | Cysteine-rich repeat secretory-like protein | Up |
| AT4G20530 | Cysteine-rich repeat secretory-like protein | Up |
| AT3G22000 | Cysteine-rich repeat secretory protein, putative (DUF26) | Up |
| AT3G21920 | Cysteine-rich repeat secretory protein, putative (DUF26) | Up |
| AT3G29040 | Cysteine-rich repeat secretory protein, putative (DUF26) | Up |
| AT4G20530 | Cysteine-rich repeat secretory-like protein | Up |
| AT4G20530 | Cysteine-rich repeat secretory-like protein | Up |
| AT3G21945 | Cysteine-rich repeat secretory protein | Up |
| AT3G22050 | Cysteine-rich repeat secretory protein, putative (DUF26) | Up |
| AT4G20680 | Cysteine-rich repeat secretory protein, putative (DUF26) | Up |
| AT3G21990 | Cysteine-rich repeat secretory-like protein (DUF26) | Up |
| LCR5 | Low-molecular-weight cysteine-rich 5 | Up |
| LCR1 | Low-molecular-weight cysteine-rich 1 | Up |
| LCR33 | Low-molecular-weight cysteine-rich 33 | Up |
| LCR4 | Low-molecular-weight cysteine-rich 4 | Up |
| LCR32 | Low-molecular-weight cysteine-rich 32 | Up |
| LCR25 | Low-molecular-weight cysteine-rich 25 | Up |
| LCR10 | Low-molecular-weight cysteine-rich 10 | Up |
| AT2G33690 | Late embryosis abundant protein, group 6 | Up |
| AT4G13230 | Late embryosis abundant protein (LEA) family protein | Up |
| YLS9 | Late embryosis abundant (LEA) hydroxyproline-rich glycoprotein family | Up |
| AT1G15415 | Late embryosis abundant-like protein | Up |
| AT5G53820 | Late embryosis abundant protein (LEA) family protein | Up |
| AT2G38390 | Peroxidase superfamily protein | Down |
| AT5G64110 | Peroxidase superfamily protein | Down |
| AT2G38380 | Peroxidase superfamily protein | Down |
| AT3G32980 | Peroxidase superfamily protein | Down |
| AT2G35380 | Peroxidase superfamily protein | Down |
| AT1G68850 | Peroxidase superfamily protein | Down |
| AT2G18150 | Peroxidase superfamily protein | Down |
| AT3G21770 | Peroxidase superfamily protein | Down |
| AT4G36430 | Peroxidase superfamily protein | Down |
| AT5G58390 | Peroxidase superfamily protein | Down |
| AT5G06730 | Peroxidase superfamily protein | Down |
| PRXR1 | Peroxidase superfamily protein | Down |
| CYP702A6 | Cytochrome P450, family 702, subfamily A, polypeptide 6 | Down |
| CYP81F4 | Cytochrome P450, family 81, subfamily F, polypeptide 4 | Down |
| CYP708A2 | Cytochrome P450, family 708, subfamily A, polypeptide 2 | Down |
| CYP71A19 | Cytochrome P450, family 71, subfamily A, polypeptide 19 | Down |
| CYP86A1 | Cytochrome P450, family 86, subfamily A, polypeptide 1 | Down |
| CYP86B1 | Cytochrome P450, family 86, subfamily B, polypeptide 1 | Down |
| CYP72A14 | Cytochrome P450, family 72, subfamily A, polypeptide 14 | Down |
| AT1G50060 | CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathosis-related 1 protein) superfamily protein | Down |
| AT4G33710 | CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathosis-related 1 protein) superfamily protein | Down |
| AT4G33720 | CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathosis-related 1 protein) superfamily protein | Down |
| AT2G19970 | CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathosis-related 1 protein) superfamily protein | Down |
| AT1G52100 | Mannose-binding lectin superfamily protein | Down |
| AT5G38550 | Mannose-binding lectin superfamily protein | Down |
| JAL33 | Mannose-binding lectin superfamily protein | Down |
| AT1G52120 | Mannose-binding lectin superfamily protein | Down |
| AT1G52000 | Mannose-binding lectin superfamily protein | Down |
| AT5G49850 | Mannose-binding lectin superfamily protein | Down |
表2 上下调基因中差异显著且富集的主要基因统计表
Table 2 Statistical table of key genes with significant differential expression and enrichment in up and down genes
基因名 Gene name | 基因描述 Gene description | 调控 Regulate |
|---|---|---|
| AT1G71680 | Transmembrane amino acid transporter family protein | Up |
| STP6 | Sugar transporter 6 | Up |
| PMT2 | Polyol/monosaccharide transporter 2 | Up |
| ACA7 | Cation transporter/E1-E2 ATPase family protein | Up |
| STP9 | Sugar transporter 9 | Up |
| CAT7 | Cationic amino acid transporter 7 | Up |
| AST91 | Sulfate transporter 91 | Up |
| PHT3;2 | Phosphate transporter 3;2 | Up |
| LHT1 | Lysine histidine transporter 1 | Up |
| ABCG1 | ABC-2 type transporter family protein | Up |
| GA20OX3 | Gibberellin 20-oxidase 3 | Up |
| GA20OX2 | Gibberellin 20 oxidase 2 | Up |
| GA3OX1 | Gibberellin 3-oxidase 1 | Up |
| AT3G21970 | Cysteine-rich repeat secretory protein, putative (DUF26) | Up |
| AT4G20530 | Cysteine-rich repeat secretory-like protein | Up |
| AT4G20530 | Cysteine-rich repeat secretory-like protein | Up |
| AT4G20530 | Cysteine-rich repeat secretory-like protein | Up |
| AT4G20530 | Cysteine-rich repeat secretory-like protein | Up |
| AT3G22000 | Cysteine-rich repeat secretory protein, putative (DUF26) | Up |
| AT3G21920 | Cysteine-rich repeat secretory protein, putative (DUF26) | Up |
| AT3G29040 | Cysteine-rich repeat secretory protein, putative (DUF26) | Up |
| AT4G20530 | Cysteine-rich repeat secretory-like protein | Up |
| AT4G20530 | Cysteine-rich repeat secretory-like protein | Up |
| AT3G21945 | Cysteine-rich repeat secretory protein | Up |
| AT3G22050 | Cysteine-rich repeat secretory protein, putative (DUF26) | Up |
| AT4G20680 | Cysteine-rich repeat secretory protein, putative (DUF26) | Up |
| AT3G21990 | Cysteine-rich repeat secretory-like protein (DUF26) | Up |
| LCR5 | Low-molecular-weight cysteine-rich 5 | Up |
| LCR1 | Low-molecular-weight cysteine-rich 1 | Up |
| LCR33 | Low-molecular-weight cysteine-rich 33 | Up |
| LCR4 | Low-molecular-weight cysteine-rich 4 | Up |
| LCR32 | Low-molecular-weight cysteine-rich 32 | Up |
| LCR25 | Low-molecular-weight cysteine-rich 25 | Up |
| LCR10 | Low-molecular-weight cysteine-rich 10 | Up |
| AT2G33690 | Late embryosis abundant protein, group 6 | Up |
| AT4G13230 | Late embryosis abundant protein (LEA) family protein | Up |
| YLS9 | Late embryosis abundant (LEA) hydroxyproline-rich glycoprotein family | Up |
| AT1G15415 | Late embryosis abundant-like protein | Up |
| AT5G53820 | Late embryosis abundant protein (LEA) family protein | Up |
| AT2G38390 | Peroxidase superfamily protein | Down |
| AT5G64110 | Peroxidase superfamily protein | Down |
| AT2G38380 | Peroxidase superfamily protein | Down |
| AT3G32980 | Peroxidase superfamily protein | Down |
| AT2G35380 | Peroxidase superfamily protein | Down |
| AT1G68850 | Peroxidase superfamily protein | Down |
| AT2G18150 | Peroxidase superfamily protein | Down |
| AT3G21770 | Peroxidase superfamily protein | Down |
| AT4G36430 | Peroxidase superfamily protein | Down |
| AT5G58390 | Peroxidase superfamily protein | Down |
| AT5G06730 | Peroxidase superfamily protein | Down |
| PRXR1 | Peroxidase superfamily protein | Down |
| CYP702A6 | Cytochrome P450, family 702, subfamily A, polypeptide 6 | Down |
| CYP81F4 | Cytochrome P450, family 81, subfamily F, polypeptide 4 | Down |
| CYP708A2 | Cytochrome P450, family 708, subfamily A, polypeptide 2 | Down |
| CYP71A19 | Cytochrome P450, family 71, subfamily A, polypeptide 19 | Down |
| CYP86A1 | Cytochrome P450, family 86, subfamily A, polypeptide 1 | Down |
| CYP86B1 | Cytochrome P450, family 86, subfamily B, polypeptide 1 | Down |
| CYP72A14 | Cytochrome P450, family 72, subfamily A, polypeptide 14 | Down |
| AT1G50060 | CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathosis-related 1 protein) superfamily protein | Down |
| AT4G33710 | CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathosis-related 1 protein) superfamily protein | Down |
| AT4G33720 | CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathosis-related 1 protein) superfamily protein | Down |
| AT2G19970 | CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathosis-related 1 protein) superfamily protein | Down |
| AT1G52100 | Mannose-binding lectin superfamily protein | Down |
| AT5G38550 | Mannose-binding lectin superfamily protein | Down |
| JAL33 | Mannose-binding lectin superfamily protein | Down |
| AT1G52120 | Mannose-binding lectin superfamily protein | Down |
| AT1G52000 | Mannose-binding lectin superfamily protein | Down |
| AT5G49850 | Mannose-binding lectin superfamily protein | Down |
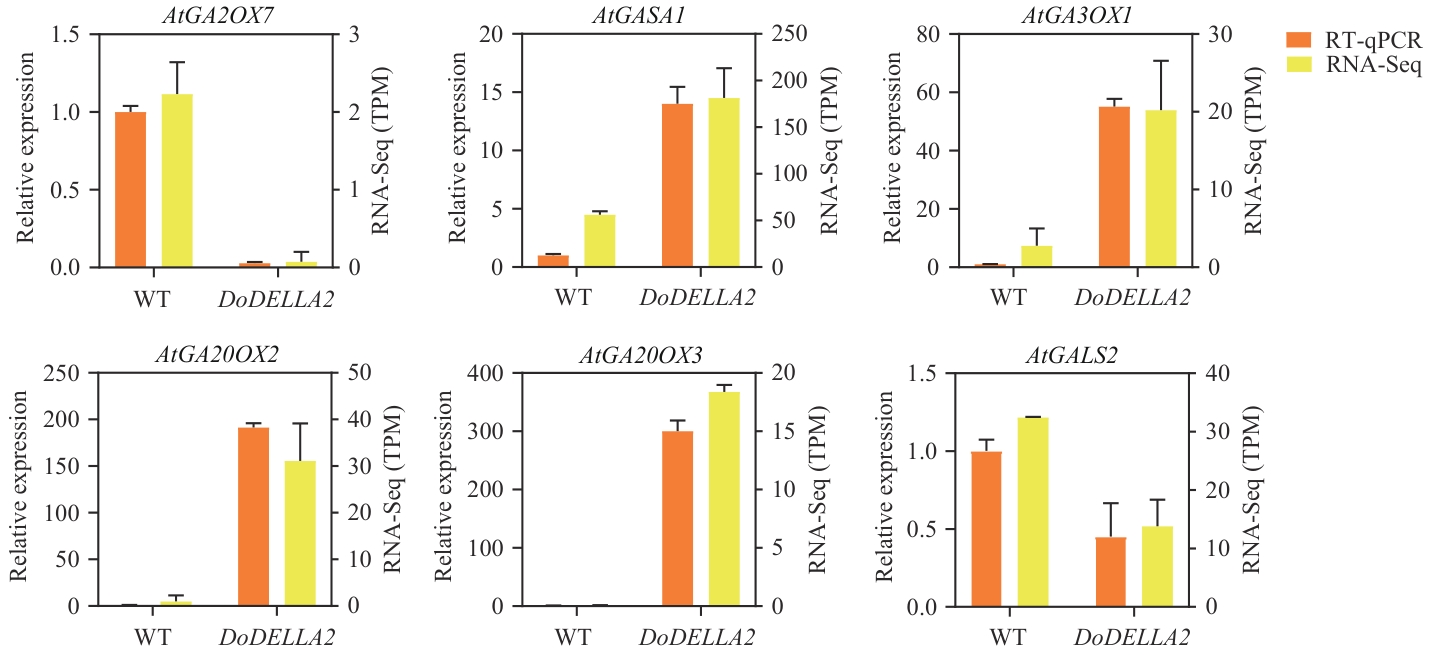
图10 赤霉素信号通路相关差异基因RT-qPCR表达水平验证WT:野生型拟南芥;DoDELLA2:过表达DoDELLA2的拟南芥
Fig. 10 Validation of RT-qPCR expression levels of differentially expressed genes related to the gibberellin signaling pathwayWT: Wild-type A. thaliana. DoDELLA2: A. thaliana overexpressing DoDELLA2
| [1] | Reinecke DM, Wickramarathna AD, Ozga JA, et al. Gibberellin 3-oxidase gene expression patterns influence gibberellin biosynthesis, growth, and development in pea [J]. Plant Physiol, 2013, 163(2): 929-945. |
| [2] | Eriksson S, Böhlenius H, Moritz T, et al. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation [J]. Plant Cell, 2006, 18(9): 2172-2181. |
| [3] | Webster AD. Vigour mechanisms in dwarfing rootstocks for temperate fruit trees [J]. Acta Hortic, 2004(658): 29-41. |
| [4] | Peng JR, Harberd NP. The role of GA-mediated signalling in the control of seed germination [J]. Curr Opin Plant Biol, 2002, 5(5): 376-381. |
| [5] | Zhang N, Xie YD, Guo HJ, et al. Gibberellins regulate the stem elongation rate without affecting the mature plant height of a quick development mutant of winter wheat (Triticum aestivum L.) [J]. Plant Physiol Biochem, 2016, 107: 228-236. |
| [6] | Wang GL, Que F, Xu ZS, et al. Exogenous gibberellin altered morphology, anatomic and transcriptional regulatory networks of hormones in carrot root and shoot [J]. BMC Plant Biol, 2015, 15: 290. |
| [7] | Appleford NE, Lenton JR. Gibberellins and leaf expansion in near-isogenic wheat lines containing Rht1 and Rht3 dwarfing alleles [J]. Planta, 1991, 183(2): 229-236. |
| [8] | Giacomelli L, Rota-Stabelli O, Masuero D, et al. Gibberellin metabolism in Vitis vinifera L. during bloom and fruit-set: functional characterization and evolution of grapevine gibberellin oxidases [J]. J Exp Bot, 2013, 64(14): 4403-4419. |
| [9] | Chen S, Wang XJ, Zhang LY, et al. Identification and characterization of tomato gibberellin 2-oxidases (GA2oxs) and effects of fruit-specific SlGA2ox1 overexpression on fruit and seed growth and development [J]. Hortic Res, 2016, 3: 16059. |
| [10] | Song PL, Li G, Xu JF, et al. Genome-wide analysis of genes involved in the GA signal transduction pathway in ‘Duli’ pear (Pyrus betulifolia Bunge) [J]. Int J Mol Sci, 2022, 23(12): 6570. |
| [11] | Achard P, Gusti A, Cheminant S, et al. Gibberellin signaling controls cell proliferation rate in Arabidopsis [J]. Curr Biol, 2009, 19(14): 1188-1193. |
| [12] | Wang Y, He SE, Wei YZ, et al. Molecular and functional characterization of two DELLA protein-coding genes in Litchi [J]. Gene, 2020, 738: 144455. |
| [13] | 王倩. 百合LiDELLA基因的克隆及初步功能分析 [D]. 长春: 吉林农业大学, 2019. |
| Wang Q. Cloning and preliminary functional analysis of LiDELLA gene from Lilium [D]. Changchun: Jilin Agricultural University, 2019. | |
| [14] | 李露露. 谷子DELLA基因的鉴定、克隆与功能分析 [D]. 太谷: 山西农业大学, 2022. |
| Li LL. Identification, cloning and functional analysis of DELLA gene in Setaria italica [D]. Taigu: Shanxi Agricultural University, 2022. | |
| [15] | 李翠. 花生DELLA家族基因的表达及转基因研究 [D]. 广州: 仲恺农业工程学院, 2015. |
| Li C. Expression and transgenic study of DELLA family genes in peanut [D]. Guangzhou: Zhongkai University of Agriculture and Engineering, 2015. | |
| [16] | Liu Q, Wu K, Harberd NP, et al. Green revolution DELLAs: From translational reinitiation to future sustainable agriculture [J]. Mol Plant, 2021, 14(4): 547-549. |
| [17] | 倪凯, 何鹏飞, 梁志庆, 等. 铁皮石斛化学成分、药理作用及毒理学评价研究进展 [J]. 云南中医中药杂志, 2023, 44(10): 86-93. |
| Ni K, He PF, Liang ZQ, et al. Research progress on chemical constituents, pharmacological effects and toxicological evaluation of Dendrobium candidum [J]. Yunnan J Tradit Chin Med Mater Med, 2023, 44(10): 86-93. | |
| [18] | 马庆, 付强, 邹西西, 等. 铁皮石斛DELLA家族基因克隆与生物信息学分析 [J]. 贵州中医药大学学报, 2022, 44(6): 13-19. |
| Ma Q, Fu Q, Zou XX, et al. Cloning and bioinformatics analysis of DELLA family gene of Dendrobium candidum [J]. J Guizhou Univ Tradit Chin Med, 2022, 44(6): 13-19. | |
| [19] | 李琳, 付强, 杨涛, 等. 铁皮石斛种子cDNA酵母文库的构建及DELLA互作蛋白筛选 [J]. 植物科学学报, 2025, 43(2): 221-229. |
| Li L, Fu Q, Yang T, et al. cDNA yeast library construction of Dendrobium officinale Kimura et Migo seeds and screening and analysis of DELLA interacting proteins [J]. Plant Sci J, 2025, 43(2): 221-229. | |
| [20] | 王健, 杨莎, 孙庆文, 等. 金钗石斛bHLH转录因子家族全基因组鉴定及表达分析 [J]. 生物技术通报, 2024, 40(6): 203-218. |
| Wang J, Yang S, Sun QW, et al. Genome-wide identification and expression analysis of bHLH transcription factor family in Dendrobium nobile [J]. Biotechnol Bull, 2024, 40(6): 203-218. | |
| [21] | 黄龙成. 拟南芥PTED1基因影响花粉壁模式形成的机理研究 [D]. 上海: 上海师范大学, 2025. |
| Huang LC. Study on the mechanism of PTED1 gene influencing pollen wall pattern formation in Arabidopsis thaliana [D]. Shanghai: Shanghai Normal University, 2025. | |
| [22] | 陈紫竹, 庆军, 格根塔娜, 等. 基于转录组测序的枣胚败育关键基因挖掘 [J]. 种子, 2025, 44(2): 95-104. |
| Chen ZZ, Qing J, Ge G, et al. Extraction of key genes of Ziziphus jujuba embryo abortion based on transcriptome sequencing [J]. Seed, 2025, 44(2): 95-104. | |
| [23] | 陈凯, 王灏, 陈燚婷, 等. 铁皮石斛WOX家族基因在生长发育中的功能分析 [J]. 遗传, 2023, 45(8): 700-714. |
| Chen K, Wang H, Chen YT, et al. Functional analysis of WOX family genes in Dendrobium catenatum during growth and development [J]. Hereditas: Beijing, 2023, 45(8): 700-714. | |
| [24] | Wilson RN, Somerville CR. Phenotypic suppression of the gibberellin-insensitive mutant (Gai) of Arabidopsis [J]. Plant Physiol, 1995, 108(2): 495-502. |
| [25] | Silverstone AL, Ciampaglio CN, Sun T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway [J]. Plant Cell, 1998, 10(2): 155-169. |
| [26] | 赵静一, 吴小旭, 胡云捷, 等. 洋葱AcGAI的克隆及其在开花途径的功能分析 [J]. 园艺学报, 2024, 51(8): 1792-1802. |
| Zhao JY, Wu XX, Hu YJ, et al. Molecular cloning and functional analysis of AcGAI in onion flowering regulation [J]. Acta Hortic Sin, 2024, 51(8): 1792-1802. | |
| [27] | 贾玉凤. 小麦过氧化物酶家族基因功能分析及小麦矮腥黑粉菌效应蛋白研究 [D]. 乌鲁木齐: 新疆农业大学, 2024. |
| Jia YF. Functional analysis of the wheat peroxidase gene family and research on tilletia controversa Kühn effector proteins [D]. Urumqi: Xinjiang Agricultural University, 2024. | |
| [28] | 钟春梅, 王小菁. 富含半胱氨酸的GASA小分子蛋白研究进展 [J]. 植物学报, 2016, 51(1): 1-8. |
| Zhong CM, Wang XJ. Progress in cysteine-rich gibberellic acid-stimulated Arabidopsis protein [J]. Chin Bull Bot, 2016, 51(1): 1-8. | |
| [29] | Khalifa MAS, Zhang Q, Du YY, et al. Functional characterisation of GmGASA1-like gene in Glycine max (L.) Merr. overexpression promotes growth, development and stress responses [J]. Life, 2024, 14(11): 1436. |
| [30] | 宋建播. 拟南芥局部及系统获得抗性的脂质代谢组学研究及GASA1基因的抗病功能验证 [D]. 西安: 西北大学, 2022. |
| Song JB. Lipid metabonomics study on local and systemic acquired resistance in Arabidopsis thaliana and verification of disease resistance function of GASA1 gene [D]. Xi’an: Northwest University, 2022. | |
| [31] | Wang L, Ma CR, Wang SH, et al. Ethylene and jasmonate signaling converge on gibberellin catabolism during thigmomorphogenesis in Arabidopsis [J]. Plant Physiol, 2024, 194(2): 758-773. |
| [32] | Gao XM, Lou SL, Han Y, et al. Allelic variations in GA20ox3 regulate fruit length and seed germination timing for high-altitude adaptation in Arabidopsis thaliana [J]. Nat Commun, 2025, 16(1): 5053. |
| [33] | Paciorek T, Chiapelli BJ, Wang JY, et al. Targeted suppression of gibberellin biosynthetic genes ZmGA20ox3 and ZmGA20ox5 produces a short stature maize ideotype [J]. Plant Biotechnol J, 2022, 20(6): 1140-1153. |
| [34] | Cheng JY, Hill C, Han Y, et al. New semi-dwarfing alleles with increased coleoptile length by gene editing of gibberellin 3-oxidase 1 using CRISPR-Cas9 in barley (Hordeum vulgare L.) [J]. Plant Biotechnol J, 2023, 21(4): 806-818. |
| [35] | Chakraborty P, Biswas A, Dey S, et al. Cytochrome P450 gene families: role in plant secondary metabolites production and plant defense [J]. J Xenobiot, 2023, 13(3): 402-423. |
| [36] | Tan HJ, Man C, Xie Y, et al. A crucial role of GA-regulated flavonol biosynthesis in root growth of Arabidopsis [J]. Mol Plant, 2019, 12(4): 521-537. |
| [37] | Xue HD, Gao X, He P, et al. Origin, evolution, and molecular function of DELLA proteins in plants [J]. Crop J, 2022, 10(2): 287-299. |
| [38] | Achard P, Cheng H, De Grauwe L, et al. Integration of plant responses to environmentally activated phytohormonal signals [J]. Science, 2006, 311(5757): 91-94. |
| [1] | 王芳, 邵会茹, 吕林龙, 赵点, 胡振, 吕建珍, 姜亮. 植物和细菌TurboID邻近蛋白标记方法的建立[J]. 生物技术通报, 2025, 41(9): 44-53. |
| [2] | 刘建国, 刘格儿, 郭颖欣, 王斌, 王玉昆, 卢金凤, 黄文庭, 朱云娜. 转录组和代谢组联合解析‘桂柚1号’和‘沙田柚’果实品质差异[J]. 生物技术通报, 2025, 41(9): 168-181. |
| [3] | 刘泽洲, 段乃彬, 岳丽昕, 王清华, 姚行浩, 高莉敏, 孔素萍. 大蒜叶片蜡质成分分析及蜡质缺失基因Ggl-1筛选[J]. 生物技术通报, 2025, 41(9): 219-231. |
| [4] | 闫梦阳, 梁晓阳, 戴君昂, 张妍, 关团, 张辉, 刘良波, 孙志华. 阿莫西林降解菌的筛选及降解机制研究[J]. 生物技术通报, 2025, 41(9): 314-325. |
| [5] | 刘佳丽, 宋经荣, 赵文宇, 张馨元, 赵子洋, 曹一博, 张凌云. 蓝莓R2R3-MYB基因鉴定及类黄酮调控基因表达分析[J]. 生物技术通报, 2025, 41(9): 124-138. |
| [6] | 李玉珍, 李梦丹, 张蔚, 彭婷. 基于月季扩展蛋白基因家族鉴定的野蔷薇RmEXPB2基因功能研究[J]. 生物技术通报, 2025, 41(9): 182-194. |
| [7] | 白雨果, 李婉迪, 梁建萍, 石志勇, 卢庚龙, 刘红军, 牛景萍. 哈茨木霉T9131对黄芪幼苗的促生机理[J]. 生物技术通报, 2025, 41(8): 175-185. |
| [8] | 李雅琼, 格桑拉毛, 陈启迪, 杨宇环, 何花转, 赵耀飞. 异源过表达高粱SbSnRK2.1增强拟南芥对盐胁迫的抗性[J]. 生物技术通报, 2025, 41(8): 115-123. |
| [9] | 曾丹, 黄园, 王健, 张艳, 刘庆霞, 谷荣辉, 孙庆文, 陈宏宇. 铁皮石斛bZIP转录因子家族全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 197-210. |
| [10] | 柴军发, 洪波, 贾彦霞. 转录组和代谢组联合分析三株蜡蚧轮枝菌菌株毒力差异[J]. 生物技术通报, 2025, 41(8): 311-321. |
| [11] | 王月琛, 韩鑫骐, 魏文敏, 崔兆兰, 罗阳美, 陈鹏如, 王海岗, 刘龙龙, 张莉, 王纶. 黍稷落粒的生物学基础研究及落粒调控基因的鉴定[J]. 生物技术通报, 2025, 41(7): 164-171. |
| [12] | 黄旭升, 周雅莉, 柴旭东, 闻婧, 王计平, 贾小云, 李润植. 紫苏质体型PfLPAT1B基因的克隆及其在油脂合成中的功能分析[J]. 生物技术通报, 2025, 41(7): 226-236. |
| [13] | 张越, 毕钰, 慕雪男, 郑子薇, 王志刚, 徐伟慧. 小麦赤霉病拮抗菌JB7的生防特性[J]. 生物技术通报, 2025, 41(7): 261-271. |
| [14] | 李成花, 豆飞飞, 任毓昭, 刘彩霞, 刘凤楼, 王掌军, 李清峰. 外施水杨酸对白粉菌侵染小麦的影响及白粉菌转录组分析[J]. 生物技术通报, 2025, 41(7): 272-280. |
| [15] | 郭秀娟, 冯宇, 吴瑞香, 王利琴, 杨建春. Ca2+处理对胡麻种子萌发影响的转录组分析[J]. 生物技术通报, 2025, 41(7): 139-149. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||