








生物技术通报 ›› 2020, Vol. 36 ›› Issue (11): 245-258.doi: 10.13560/j.cnki.biotech.bull.1985.2019-1273
王琦1( ), 颜春蕾1, 高洪伟2, 吴薇1(
), 颜春蕾1, 高洪伟2, 吴薇1( ), 杨庆利1(
), 杨庆利1( )
)
收稿日期:2019-12-29
出版日期:2020-11-26
发布日期:2020-11-20
作者简介:王琦,女,硕士研究生,研究方向:食品危害物的快速检测;E-mail: 基金资助:
WANG Qi1( ), YAN Chun-lei1, GAO Hong-wei2, WU Wei1(
), YAN Chun-lei1, GAO Hong-wei2, WU Wei1( ), YANG Qing-li1(
), YANG Qing-li1( )
)
Received:2019-12-29
Published:2020-11-26
Online:2020-11-20
摘要:
核酸适配体是可以识别并与靶标分子结合的单链DNA或RNA分子。由于适配体具有良好的特异性、稳定性、选择性、易于修饰和亲和力高等特点,在病原检测和其他小分子识别方面具有很大的潜力。综述了适配体的筛选技术以及适配体应用技术,包括比色法、可见光谱法、荧光法、电化学法、表面增强拉曼法和胶体金试纸条法等在食源性致病菌检测中的应用。目前适配体传感器在检测食品致病菌领域取得了显著进展。然而核酸适配体应用方面的挑战和缺陷仍需克服。
王琦, 颜春蕾, 高洪伟, 吴薇, 杨庆利. 基于核酸适配体传感器检测食品致病菌的研究进展[J]. 生物技术通报, 2020, 36(11): 245-258.
WANG Qi, YAN Chun-lei, GAO Hong-wei, WU Wei, YANG Qing-li. Research Progress of DNA Aptasensors for Foodborne Pathogen Detection[J]. Biotechnology Bulletin, 2020, 36(11): 245-258.
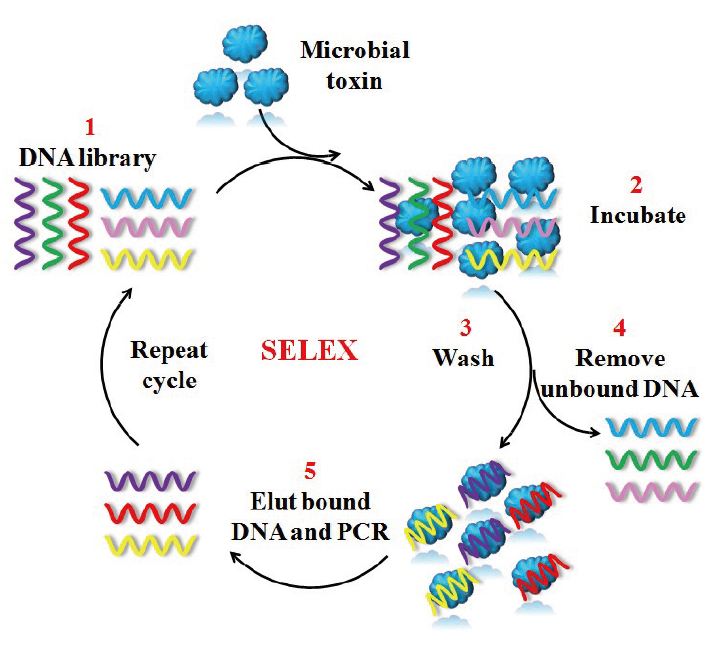
图1 利用SELEX筛选食源性致病菌适配体的示意图 SELEX分为5个步骤:1. 合成体外DNA随机文库;2. DNA文库和靶标病原菌一起孵育;3. 未结合核酸序列的分离;4. 与靶标结合的核酸序列的洗脱;5. 目标结合核酸序列作为下一次PCR扩增的模板。扩增后的产物作为新的DNA文库用于下一轮的筛选
| 靶标 | 菌型 | 筛选方法 | 序列(5'-3') | 参考文献 |
|---|---|---|---|---|
| 大肠杆菌 | 大肠杆菌O157:H7 | 全细胞-SELEX | ACCAGTAGACTTTCAACTTTACTGCCATCGTGTGCCCTAA | [25,32] |
| Non-SELEX | TATGGCGTGGCAAGCTTGGCCCGCTTCTCAAGCATGGTTATCTAC | [33] | ||
| 大肠杆菌O111:B4 | 磁珠-SELEX | ATCCGTCACCCCTGCTCTCGTCGCTATGAAGTAACAAAGATAGGAGCAAT CGGGTGGTGTTGGCTCCCGTAT | [34] | |
| 大肠杆菌O111 | 全细胞-SELEX | ATCCGTCACACCTGCTCTACTGGCCGGCTCAGCATGACTAAGAAGGAAGT TATGTGGTGTTGGCTCCCGTAT | [35] | |
| 大肠杆菌O55:B5 | Non-SELEX | TAGCCGGATCGCGCTGGCCAGATGATATAAAGGGTCAGCCCCCCA | [36] | |
| 沙门氏菌 | 鼠伤寒沙门氏菌 | 全细胞-SELEX | ACGGGCGTGGGGGCAATGCCTGCTTGTAGGCTTCCCCTGTGCGCG | [37] |
| SELEX | TATGGCGGCGTCACCCGACGGGGACTTGACATTATGACAG | [38] | ||
| 沙门氏菌O8 | 全细胞-SELEX | GATCCGGGCCTCATGTCGAACACCCCCCAACTAAAACAACAAAACACC ACCGCCATTGAGCGTTTATTCTGAGCTCCCA | [39] | |
| SELEX | TGATCCGGGCCTCATGTCGAACACCCCCCAACTAAAACAACAAAACAC CACCGCCATTGAGCGTTTATTCTGGCTCCCA | [40] | ||
| 肠炎沙门氏菌 | 磁珠-SELEX | GGGUUCACUGCAGACUUGACGAAGCUUGAGAGAUGCCCCCUGAUGTGC AUUCUUGUUGUGUUGCGGCAAUGGAUCCACAUCTACGAAUUC | [41] | |
| 金黄色葡萄球菌 | 金黄色葡萄球菌 | 全细胞-SELEX | GCGCCCTCTCACGTGGCACTCAGAGTGCCGGAAGTTCTGCGTTAT | [42] |
| 肠毒素 | SELEX | TTTGGTATTGAGGGTCGCATCCACTGGTCGTTGTCTGTTGTCTGTTAT GTTGTTTCGTGATGGCTCTAACTCTCCTCT | [43] | |
| 磷壁酸 | SELEX | GGGAGUUUUGAUACGGCUUCAUGCAGUAAUGUUUUUAU | [31] | |
| 肠毒素B | SELEX | AGCAGCACAGAGGTCAGATGTATACTTCTAAAATTTGTTTGTATCTAC GATGTTCTTCGTCCTATGCGTGCTACCGTGAA | [44] | |
| 肠毒素A | SELEX | TACTTATGCATTTCCTCCCACGATCTTATTTGAGAGTGAC | [45] | |
| 副溶血弧菌 | 全细胞-SELEX | ATAGGAGTCACGACGACCAGAATCTAAAAATGGGCAAAGAAACAGT GACTCGTTGAGATACTTATGTGCGTCTACCTCTTGACTAAT | [46] | |
| 单核细胞增生李斯特菌 | 全细胞-SELEX | TGGGAGCTCAGAATAAACGCTCAACTTTGTTCTTCTTTGCTTTTTTTTT CTTTTTTTGTTCGACATGAGGCCCGGATCA | [47] | |
| 蜡样芽孢杆菌 | 芽孢 | SELEX | CATCCGTCACACCTGCTCGGTGCAGACCCATAGGGGGGGCGTGCGGA TGTAGGAGTAGGGTGTTGGCTCCCGTATC | [48] |
| 志贺氏菌 | 痢疾志贺氏菌 | 全细胞-SELEX | CGGAACTAGCGTTTAAATGCCAGGACTGAAGTAGGCAGGG | [22] |
| 宋内志贺菌 | 全细胞-SELEX | ATTATGTTGTCTGAACTGCCATGGTCCCTCGTGTTTATTATGTTGTCT GACTGGCTGAGATTGCACTTACTATCT | [49] | |
| 绿脓杆菌 | 全细胞-SELEX | CCCCCGTTGCTTTCGCTTTTCCTTTCGCTTTTGTTCGTTTCGTCCCTGC TTCCTTTCTTG | [50] |
表1 不同筛选方法得到的食源性致病菌的适配体
| 靶标 | 菌型 | 筛选方法 | 序列(5'-3') | 参考文献 |
|---|---|---|---|---|
| 大肠杆菌 | 大肠杆菌O157:H7 | 全细胞-SELEX | ACCAGTAGACTTTCAACTTTACTGCCATCGTGTGCCCTAA | [25,32] |
| Non-SELEX | TATGGCGTGGCAAGCTTGGCCCGCTTCTCAAGCATGGTTATCTAC | [33] | ||
| 大肠杆菌O111:B4 | 磁珠-SELEX | ATCCGTCACCCCTGCTCTCGTCGCTATGAAGTAACAAAGATAGGAGCAAT CGGGTGGTGTTGGCTCCCGTAT | [34] | |
| 大肠杆菌O111 | 全细胞-SELEX | ATCCGTCACACCTGCTCTACTGGCCGGCTCAGCATGACTAAGAAGGAAGT TATGTGGTGTTGGCTCCCGTAT | [35] | |
| 大肠杆菌O55:B5 | Non-SELEX | TAGCCGGATCGCGCTGGCCAGATGATATAAAGGGTCAGCCCCCCA | [36] | |
| 沙门氏菌 | 鼠伤寒沙门氏菌 | 全细胞-SELEX | ACGGGCGTGGGGGCAATGCCTGCTTGTAGGCTTCCCCTGTGCGCG | [37] |
| SELEX | TATGGCGGCGTCACCCGACGGGGACTTGACATTATGACAG | [38] | ||
| 沙门氏菌O8 | 全细胞-SELEX | GATCCGGGCCTCATGTCGAACACCCCCCAACTAAAACAACAAAACACC ACCGCCATTGAGCGTTTATTCTGAGCTCCCA | [39] | |
| SELEX | TGATCCGGGCCTCATGTCGAACACCCCCCAACTAAAACAACAAAACAC CACCGCCATTGAGCGTTTATTCTGGCTCCCA | [40] | ||
| 肠炎沙门氏菌 | 磁珠-SELEX | GGGUUCACUGCAGACUUGACGAAGCUUGAGAGAUGCCCCCUGAUGTGC AUUCUUGUUGUGUUGCGGCAAUGGAUCCACAUCTACGAAUUC | [41] | |
| 金黄色葡萄球菌 | 金黄色葡萄球菌 | 全细胞-SELEX | GCGCCCTCTCACGTGGCACTCAGAGTGCCGGAAGTTCTGCGTTAT | [42] |
| 肠毒素 | SELEX | TTTGGTATTGAGGGTCGCATCCACTGGTCGTTGTCTGTTGTCTGTTAT GTTGTTTCGTGATGGCTCTAACTCTCCTCT | [43] | |
| 磷壁酸 | SELEX | GGGAGUUUUGAUACGGCUUCAUGCAGUAAUGUUUUUAU | [31] | |
| 肠毒素B | SELEX | AGCAGCACAGAGGTCAGATGTATACTTCTAAAATTTGTTTGTATCTAC GATGTTCTTCGTCCTATGCGTGCTACCGTGAA | [44] | |
| 肠毒素A | SELEX | TACTTATGCATTTCCTCCCACGATCTTATTTGAGAGTGAC | [45] | |
| 副溶血弧菌 | 全细胞-SELEX | ATAGGAGTCACGACGACCAGAATCTAAAAATGGGCAAAGAAACAGT GACTCGTTGAGATACTTATGTGCGTCTACCTCTTGACTAAT | [46] | |
| 单核细胞增生李斯特菌 | 全细胞-SELEX | TGGGAGCTCAGAATAAACGCTCAACTTTGTTCTTCTTTGCTTTTTTTTT CTTTTTTTGTTCGACATGAGGCCCGGATCA | [47] | |
| 蜡样芽孢杆菌 | 芽孢 | SELEX | CATCCGTCACACCTGCTCGGTGCAGACCCATAGGGGGGGCGTGCGGA TGTAGGAGTAGGGTGTTGGCTCCCGTATC | [48] |
| 志贺氏菌 | 痢疾志贺氏菌 | 全细胞-SELEX | CGGAACTAGCGTTTAAATGCCAGGACTGAAGTAGGCAGGG | [22] |
| 宋内志贺菌 | 全细胞-SELEX | ATTATGTTGTCTGAACTGCCATGGTCCCTCGTGTTTATTATGTTGTCT GACTGGCTGAGATTGCACTTACTATCT | [49] | |
| 绿脓杆菌 | 全细胞-SELEX | CCCCCGTTGCTTTCGCTTTTCCTTTCGCTTTTGTTCGTTTCGTCCCTGC TTCCTTTCTTG | [50] |
| 传感器 | 适用场所 | 灵敏度CFU/mL | 优缺点 | 靶标 | LOD | 检测范围 | 参考 文献 |
|---|---|---|---|---|---|---|---|
| 比色适配体传感器 | 直观快速检测复杂的食品基质 | 10 | 结果直观,适用于现场检测,但结果难以量化 | 副溶血性弧菌 | 10 CFU/mL | 102-107CFU/mL | [57] |
| 鼠伤寒沙门氏菌 | 11 CFU/mL | 11-1.10×105CFU/mL | [58] | ||||
| 可见光适配体传感器 | POC测试与商业检测 | 100 | 检测快速、易于操作、成本低、仪器便携,但灵敏度较差,难以精确量化 | 伤寒沙门菌 | 103CFU/mL | 103-107 CFU /mL | [59] |
| 金黄色葡萄球菌 | 3.17 μmol/L | 8-23 μmol/L | [60] | ||||
| 荧光适配体传感器 | 各种高通量的样品 | 103 | 特异性高,试剂和仪器成本高 | 嗜水气单胞菌和迟缓爱德华菌 | 1.5 CFU/mL和 67 CFU/mL | 0.1-1.3×105CFU/mL和1.3-1.3×104CFU/mL | [61] |
| 电化学适配体传感器 | 痕量检测 | 金黄色葡萄球菌 | 1 CFU/mL | 10-1×106CFU/mL | [62] | ||
| O157:H7型大肠杆菌 | 10 CFU/mL | - | [56] | ||||
| 表面增强拉曼散射适配体传感器 | 痕量靶标的无损检测 | 5 | 灵敏度高,无标签,专业性要求高,仪器贵重 | 金黄色葡萄球菌 | 1.5 CFU/mL | 10-107 CFU/mL | [63] |
| 测流适配体传感器 | 直观快速检测 | 10 | 方便携带、易于操作、成本低,但难以量化 | 副溶血性弧菌 | 5.6 CFU/mL | - | [19] |
表2 用于食源性致病菌检测的传感器的性能比较
| 传感器 | 适用场所 | 灵敏度CFU/mL | 优缺点 | 靶标 | LOD | 检测范围 | 参考 文献 |
|---|---|---|---|---|---|---|---|
| 比色适配体传感器 | 直观快速检测复杂的食品基质 | 10 | 结果直观,适用于现场检测,但结果难以量化 | 副溶血性弧菌 | 10 CFU/mL | 102-107CFU/mL | [57] |
| 鼠伤寒沙门氏菌 | 11 CFU/mL | 11-1.10×105CFU/mL | [58] | ||||
| 可见光适配体传感器 | POC测试与商业检测 | 100 | 检测快速、易于操作、成本低、仪器便携,但灵敏度较差,难以精确量化 | 伤寒沙门菌 | 103CFU/mL | 103-107 CFU /mL | [59] |
| 金黄色葡萄球菌 | 3.17 μmol/L | 8-23 μmol/L | [60] | ||||
| 荧光适配体传感器 | 各种高通量的样品 | 103 | 特异性高,试剂和仪器成本高 | 嗜水气单胞菌和迟缓爱德华菌 | 1.5 CFU/mL和 67 CFU/mL | 0.1-1.3×105CFU/mL和1.3-1.3×104CFU/mL | [61] |
| 电化学适配体传感器 | 痕量检测 | 金黄色葡萄球菌 | 1 CFU/mL | 10-1×106CFU/mL | [62] | ||
| O157:H7型大肠杆菌 | 10 CFU/mL | - | [56] | ||||
| 表面增强拉曼散射适配体传感器 | 痕量靶标的无损检测 | 5 | 灵敏度高,无标签,专业性要求高,仪器贵重 | 金黄色葡萄球菌 | 1.5 CFU/mL | 10-107 CFU/mL | [63] |
| 测流适配体传感器 | 直观快速检测 | 10 | 方便携带、易于操作、成本低,但难以量化 | 副溶血性弧菌 | 5.6 CFU/mL | - | [19] |
| [1] |
Dallman TJ, Chattaway MA, Cowley LA, et al. An investigation of the diversity of strains of enteroaggregative Escherichia coli isolated from cases associated with a large multi-pathogen foodborne outbreak in the UK[J]. PLoS One, 2014,9(5):e98103.
doi: 10.1371/journal.pone.0098103 URL pmid: 24844597 |
| [2] |
Deng X, Den Bakker HC, Hendriksen RS. Genomic epidemiology:whole-genome-sequencing-powered surveillance and outbreak investigation of foodborne bacterial pathogens[J]. Annual Review of Food Science and Technology, 2016,7(1):353-374.
doi: 10.1146/annurev-food-041715-033259 URL |
| [3] |
Kirk MD, Pires SM, Black RE, et al. World health organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010:a data synjournal[J]. PLoS Medicine, 2015,12(12):e1001921.
doi: 10.1371/journal.pmed.1001921 URL pmid: 26633831 |
| [4] |
Majowicz Shannon E. Using market availability data to support foodborne disease outbreak investigations[J]. American Journal of Public Health, 2020,110(3):278-280.
doi: 10.2105/AJPH.2019.305536 URL pmid: 32023105 |
| [5] | 付萍, 刘志涛, 梁骏华, 等. 2014年中国大陆食源性疾病暴发事件监测资料分析[J]. 中国食品卫生杂志, 2018,30(6):628-634. |
| Fu P, Liu ZT, Liang JH, et al. Analysis of surveillance data of foodborne disease outbreaks in Mainland China in 2014[J]. Chinese Journal of Food Hygiene, 2018,30(6):628-634. | |
| [6] | 胡金强, 黄润娜, 王一, 等. 核酸适配体在食源性致病菌检测中应用的研究进展[J]. 食品工业科技, 2019,40(9):315-322. |
| Hu JQ, Huang RN, Wang Y, et al. Research progress on utilization of nucleic acid AptaMer in detection for foodborne pathogenic bacte[J]. Food Industry Technology, 2019,40(9):315-322. | |
| [7] |
Patel RR, Sundin GW, Yang CH, et al. Exploration of using antisense peptide nucleic acid(PNA)-cell penetrating peptide(CPP)as a novel bactericide against fire blight pathogen Erwinia amylovora[J]. Frontiers in Microbiology, 2017,8:687.
URL pmid: 28469617 |
| [8] |
Akyol I. Development and application of RTi-PCR method for common food pathogen presence and quantity in beef, sheep and chicken meat[J]. Meat Science, 2018,137:9-15.
doi: 10.1016/j.meatsci.2017.11.001 URL pmid: 29149629 |
| [9] |
Davydova A, Vorobjeva M, Pyshnyi D, et al. Aptamers against pathogenic microorganisms[J]. Critical Reviews in Microbiology, 2016,42(6):847-865.
doi: 10.3109/1040841X.2015.1070115 URL pmid: 26258445 |
| [10] |
Punnarak P, Santos MD, Hwang SD, et al. RNA aptamers inhibit the growth of the fish pathogen viral hemorrhagic Septicemia virus(VHSV)[J]. Marine Biotechnology, 2012,14(6):752-761.
URL pmid: 22527269 |
| [11] |
Acquah C, Agyei D, Obeng EM, et al. Aptamers:an emerging class of bioaffinity ligands in bioactive peptide applications[J]. Critical Reviews in Food Science and Nutrition, 2020,60(7):1195-1206.
doi: 10.1080/10408398.2018.1564234 URL pmid: 30714390 |
| [12] |
Dupont DM, Larsen N, Jensen JK, et al. Characterisation of aptamer-target interactions by branched selection and high-throughput sequencing of SELEX pools[J]. Nucleic Acids Research, 2015,43(21):e139.
doi: 10.1093/nar/gkv700 URL pmid: 26163061 |
| [13] |
Tan SY, Acquah C, Sidhu A, et al. SELEX modifications and bioanalytical techniques for aptamer-target binding characterization[J]. Critical Reviews in Analytical Chemistry, 2016,46(6):521-537.
doi: 10.1080/10408347.2016.1157014 URL pmid: 26980177 |
| [14] |
Chung J, Kang JS, Jurng JS, et al. Fast and continuous microorganism detection using aptamer-conjugated fluorescent nanoparticles on an optofluidic platform[J]. Biosensors & Bioelectronics, 2015,67:303-308.
doi: 10.1016/j.bios.2014.08.039 URL pmid: 25190089 |
| [15] |
Ling K, Jiang H, Li Y, et al. A self-assembling RNA aptamer-based graphene oxide sensor for the turn-on detection of theophylline in serum[J]. Biosensors & Bioelectronics, 2016,86:8-13.
doi: 10.1016/j.bios.2016.06.024 URL pmid: 27318104 |
| [16] |
Zou Y, Duan N, Wu S, et al. Selection, identification, and binding mechanism studies of an ssdna aptamer targeted to different stages of E-coli O157:H7[J]. Journal of Agricultural and Food Chemistry, 2018,66(22):5677-5682.
doi: 10.1021/acs.jafc.8b01006 URL pmid: 29756774 |
| [17] |
Qiao J, Meng X, Sun Y, et al. Aptamer-based fluorometric assay for direct identification of methicillin-resistant Staphylococcus aureus from clinical samples[J]. Journal of Microbiological Methods, 2018,153:92-98.
URL pmid: 30243766 |
| [18] |
Shin WR, Sekhon SS, Kim SG, et al. Aptamer-based pathogen monitoring for Salmonella enterica ser. Typhimurium[J]. Journal of Biomedical Nanotechnology, 2018,14(11):1992-2002.
doi: 10.1166/jbn.2018.2634 URL pmid: 30165934 |
| [19] |
Wu W, Zhou M, He H, et al. A sensitive aptasensor for the detection of Vibrio parahaemolyticus[J]. Sensors and Actuators B:Chemical, 2018,272:550-558.
doi: 10.1016/j.snb.2018.05.171 URL |
| [20] |
Bruno JG, Sivils JC. Further characterization and independent validation of a DNA aptamer-quantum dot-based magnetic sandwich assay for Campylobacter[J]. Folia Microbiologica, 2017,62(6):485-490.
doi: 10.1007/s12223-017-0520-0 URL pmid: 28342148 |
| [21] |
Liu H, Whitehouse CA, Lis B. Presence and persistence of salmonella in water:The impact on microbial quality of water and food safety[J]. Frontiers in Public Health, 2018,6:159.
doi: 10.3389/fpubh.2018.00159 URL pmid: 29900166 |
| [22] |
Duan N, Ding X, Wu S, et al. In vitro selection of a DNA aptamer targeted against Shigella dysenteriae[J]. Journal of Microbiological Methods, 2013,94(3):170-174.
doi: 10.1016/j.mimet.2013.06.016 URL pmid: 23811206 |
| [23] |
Dua P, Ren S, Lee SW, et al. Cell-SELEX based identification of an RNA aptamer for Escherichia coli and its use in various detection formats[J]. Molecules and Cells, 2016,39(11):807-813.
doi: 10.14348/molcells.2016.0167 URL pmid: 27871171 |
| [24] | 孙玉琼, 袁宝银, 邓美桃, 等. 核酸适配体序列优化策略的研究进展[J]. 化学传感器, 2018,38(2):12-22. |
| Sun YQ, Yuan BY, Deng MT, et al. Recent development of the optimization strategies of aptamer sequences[J]. Chemical Sensors, 2018,38(2):12-22. | |
| [25] |
Amraee M, Oloomi M, Yavari A, et al. DNA aptamer identification and characterization for E-coli O157 detection using cell based SELEX method[J]. Analytical Biochemistry, 2017,536:36-44.
doi: 10.1016/j.ab.2017.08.005 URL pmid: 28818557 |
| [26] |
Bruno JG, Richarte AM, Phillips T. Preliminary development of a DNA aptamer-magnetic bead capture electrochemiluminescence sandwich assay for brain natriuretic peptide[J]. Microchemical Journal, 2014,115:32-38.
URL pmid: 24764602 |
| [27] | Wang LL, Fang XD, Han ZQ. Existence of standing wave solutions for coupled quasilinear Schrodinger systems with critical exponents in R-N[J]. Electronic Journal of Qualitative Theory of Differential Equations, 2017(12):1-23. |
| [28] |
Dwivedi HP, Smiley RD, Jaykus LA. Selection of DNA aptamers for capture and detection of Salmonella typhimurium using a whole-cell SELEX approach in conjunction with cell sorting[J]. Applied Microbiology and Biotechnology, 2013,97(8):3677-3686.
URL pmid: 23494620 |
| [29] |
Kowalska E, Bartnicki F, Pels K, et al. The impact of immobilized metal affinity chromatography(IMAC)resins on DNA aptamer selection[J]. Analytical and Bioanalytical Chemistry, 2014,406(22):5495-5499.
doi: 10.1007/s00216-014-7937-y URL pmid: 24924211 |
| [30] |
Berezovski M, Musheev M, Drabovich A, et al. Non-SELEX selection of aptamers[J]. Journal of the American Chemical Society, 2006,128(5):1410-1411.
URL pmid: 16448086 |
| [31] |
Maeng JS, Kim N, Kim CT, et al. Rapid detection of food pathogens using RNA aptamers-immobilized slide[J]. Journal of Nanoscience and Nanotechnology, 2012,12(7):5138-5142.
doi: 10.1166/jnn.2012.6369 URL pmid: 22966534 |
| [32] |
Yu X, Chen F, Wang R, et al. Whole-bacterium SELEX of DNA aptamers for rapid detection of E. coli O157:H7 using a QCM sensor[J]. Journal of Biotechnology, 2018,266:39-49.
doi: 10.1016/j.jbiotec.2017.12.011 URL pmid: 29242148 |
| [33] |
Kim HR, Song MY, Kim BC. Rapid isolation of bacteria-specific aptamers with a non-SELEX-based method[J]. Anal Biochem, 2020,591:113542.
doi: 10.1016/j.ab.2019.113542 URL pmid: 31837967 |
| [34] |
Bruno JG, Carrillo MP, Phillips T. In vitro antibacterial effects of antilipopolysaccharide DNA aptamer-C1qrs complexes[J]. Folia Microbiologica, 2008,53(4):295-302.
doi: 10.1007/s12223-008-0046-6 URL pmid: 18759112 |
| [35] | Luo C, Lei Y, Yan L, et al. A rapid and sensitive aptamer-based electrochemical biosensor for direct detection of Escherichia coli O111[J]. Electroanalysis, 2012,24(5):1186-1191. |
| [36] |
Zhu L, Li S, Shao X, et al. Colorimetric detection and typing of E-coli lipopolysaccharides based on adual aptamer-functionalized gold nanoparticle probe[J]. Microchimica Acta, 2019,186(2):111.
doi: 10.1007/s00604-018-3212-9 URL pmid: 30637507 |
| [37] |
Moon J, Kim G, Lee S, et al. Identification of Salmonella Typhimurium-specific DNA aptamers developed using whole-cell SELEX and FACS analysis[J]. Journal of Microbiological Methods, 2013,95(2):162-166.
URL pmid: 23978634 |
| [38] |
Joshi R, Janagama H, Dwivedi HP, et al. Selection, characterization, and application of DNA aptamers for the capture and detection of Salmonella enterica serovars[J]. Molecular and Cellular Probes, 2009,23(1):20-28.
doi: 10.1016/j.mcp.2008.10.006 URL pmid: 19049862 |
| [39] |
Dwivedi HP, Smiley RD, Jaykus LA. Selection of DNA aptamers for capture and detection of Salmonella typhimurium using a whole-cell SELEX approach in conjunction with cell sorting[J]. Applied Microbiology and Biotechnology, 2013,97(8):3677-3686.
doi: 10.1007/s00253-013-4766-4 URL pmid: 23494620 |
| [40] |
Duan N, Wu S, Chen X, et al. Selection and characterization of aptamers against Salmonella typhimurium using whole-bacterium systemic evolution of ligands by exponential enrichment(SELEX)[J]. Journal of Agricultural and Food Chemistry, 2013,61(13):3229-3234.
URL pmid: 23473545 |
| [41] |
Zhang Y, Luo F, Zhang Y, et al. A sensitive assay based on specific aptamer binding for the detection of Salmonella enterica serovar Typhimurium in milk samples by microchip capillary electrophoresis[J]. Journal of Chromatography A, 2018,1534:188-194.
doi: 10.1016/j.chroma.2017.12.054 URL pmid: 29289340 |
| [42] |
Moon J, Kim G, Park SB, et al. Comparison of whole-cell SELEX methods for the identification of Staphylococcus aureus-specific DNA aptamers[J]. Sensors, 2015,15(4):8884-8897.
doi: 10.3390/s150408884 URL pmid: 25884791 |
| [43] |
Huang Y, Chen X, Duan N, et al. Selection and characterization of DNA aptamers against Staphylococcus aureus enterotoxin C1[J]. Food Chemistry, 2015,166:623-629.
doi: 10.1016/j.foodchem.2014.06.039 URL pmid: 25053102 |
| [44] |
Degrasse JA. A Single-stranded DNA aptamer that selectively binds to Staphylococcus aureus enterotoxin B[J]. PLoS One, 2012,7(3):e33410.
doi: 10.1371/journal.pone.0033410 URL pmid: 22438927 |
| [45] |
Wang K, Wu D, Chen Z, et al. Inhibition of the superantigenic activities of Staphylococcal enterotoxin A by an aptamer antagonist[J]. Toxicon, 2016,119:21-27.
doi: 10.1016/j.toxicon.2016.05.006 URL pmid: 27179422 |
| [46] |
Song S, Wang X, Xu K, et al. Selection of highly specific aptamers to Vibrio parahaemolyticus using cell-SELEX powered by functionalized graphene oxide and rolling circle amplification[J]. Analytica Chimica Acta, 2019,1052:153-162.
doi: 10.1016/j.aca.2018.11.047 URL pmid: 30685034 |
| [47] |
Suh SH, Dwivedi HP, Choi SJ, et al. Selection and characterization of DNA aptamers specific for Listeria species[J]. Analytical Biochemistry, 2014,459:39-45.
doi: 10.1016/j.ab.2014.05.006 URL pmid: 24857773 |
| [48] |
Fischer C, Huenniger T, Jarck JH, et al. Food sensing:Aptamer-based trapping of Bacillus cereus spores with specific detection via real time PCR in milk[J]. Journal of Agricultural and Food Chemistry, 2015,63(36):8050-8057.
URL pmid: 26306797 |
| [49] |
Song MS, Sekhon SS, Shin WR, et al. Detecting and discriminating Shigella sonnei using an aptamer-based fluorescent biosensor platform[J]. Molecules, 2017,22(5):825.
doi: 10.3390/molecules22050825 URL |
| [50] |
Soundy J, Day D. Selection of DNA aptamers specific for live Pseudomonas aeruginosa[J]. PLoS One, 2017,12(9):e0185385.
doi: 10.1371/journal.pone.0185385 URL pmid: 28937998 |
| [51] | 赵彤. 食源性致病菌检测现状与食品微生物危险性评估[J]. 中国卫生标准管理, 2019,10(4):7-9. |
| Zhao T. Detection status of foodborne pathogenic bacteria and food microbial risk assessment[J]. China Health Standard Management, 2019,10(4):7-9. | |
| [52] |
Poltronieri P, Primiceri E, Radhakrishnan R. EIS-based biosensors in foodborne pathogen detection with a special focus on Listeria monocytogenes[J]. Methods in Molecular Biology, 2019,1918:87-101.
URL pmid: 30580401 |
| [53] |
Li L, Li Q, Liao Z, et al. Magnetism-resolved separation and fluorescence quantification for near-simultaneous detection of multiple pathogens[J]. Analytical Chemistry, 2018,90(15):9621-9628.
doi: 10.1021/acs.analchem.8b02572 URL pmid: 30001487 |
| [54] |
Xu X, Ma X, Wang H, et al. Aptamer based SERS detection of Salmonella typhimurium using DNA-assembled gold nanodimers[J]. Microchimica Acta, 2018,185(7):325.
doi: 10.1007/s00604-018-2852-0 URL |
| [55] |
Wang L, Wang R, Chen F, et al. QCM-based aptamer selection and detection of Salmonella typhimurium[J]. Food Chemistry, 2017,221:776-782.
doi: 10.1016/j.foodchem.2016.11.104 URL pmid: 27979272 |
| [56] |
Wang L, Huang F, Cai G, et al. An electrochemical aptasensor using coaxial capillary with magnetic nanoparticle, urease catalysis and PCB electrode for rapid and sensitive detection of Escherichia coli O157:H7[J]. Nanotheranostics, 2017,1(4):403-414.
URL pmid: 29071202 |
| [57] |
Sun Y, Duan N, Ma P, et al. Colorimetric aptasensor based on truncated aptamer and trivalent dnazyme for Vibrio parahemolyticus determination[J]. Journal of Agricultural and Food Chemistry, 2019,67(8):2313-2320.
doi: 10.1021/acs.jafc.8b06893 URL pmid: 30721047 |
| [58] |
Wu S, Duan N, Qiu Y, et al. Colorimetric aptasensor for the detection of Salmonella enterica serovar typhimurium using ZnFe2O4-reduced graphene oxide nanostructures as an effective peroxidase mimetics[J]. International Journal of Food Microbiology, 2017,261:42-48.
doi: 10.1016/j.ijfoodmicro.2017.09.002 URL pmid: 28910678 |
| [59] |
Yang M, Peng Z, Ning Y, et al. Highly specific and cost-efficient detection of salmonella paratyphi a combining aptamers with single-walled carbon nanotubes[J]. Sensors, 2013,13(5):6865-6881.
doi: 10.3390/s130506865 URL pmid: 23698275 |
| [60] |
Urmann K, Reich P, Walter JG, et al. Rapid and label-free detection of protein a by aptamer-tethered porous silicon nanostructures[J]. Journal of Biotechnology, 2017,257:171-177.
URL pmid: 28131857 |
| [61] |
Zhu QY, Zhang FR, Du Y, et al. Graphene-based steganographically aptasensing system for information computing, encryption and hiding, fluorescence sensing and in vivo imaging of fish pathogens[J]. Acs Applied Materials & Interfaces, 2019,11(9):8904-8914.
doi: 10.1021/acsami.8b22592 URL pmid: 30730133 |
| [62] |
Abbaspour A, Norouz-Sarvestani F, Noon A, et al. Aptamer-conjugated silver nanoparticles for electrochemical dual-aptamer-based sandwich detection of Staphylococcus aureus[J]. Biosensors & Bioelectronics, 2015,68:149-155.
doi: 10.1016/j.bios.2014.12.040 URL pmid: 25562742 |
| [63] |
Wang C, Wang J, Li P, et al. Sonochemical synjournal of highly branched flower-like Fe3O4@SiO2@Ag microcomposites and their application as versatile SERS substrates[J]. Nanoscale, 2016,8(47):19816-19828.
doi: 10.1039/c6nr07295j URL pmid: 27878199 |
| [64] |
Alhogail S, Suaifan GARY, Zourob M. Rapid colorimetric sensing platform for the detection of Listeria monocytogenes foodborne pathogen[J]. Biosensors & Bioelectronics, 2016,86:1061-1066.
URL pmid: 27543841 |
| [65] |
Wang J, Li H, Li T, et al. Determination of bacterial DNA based on catalytic oxidation of cysteine by G-quadruplex DNAzyme generated from asymmetric PCR:Application to the colorimetric detection of Staphylococcus aureus[J]. Microchimica Acta, 2018,185(9):410.
doi: 10.1007/s00604-018-2935-y URL pmid: 30099608 |
| [66] |
Woo MA, Kim MI, Jung JH, et al. A novel colorimetric immunoassay utilizing the peroxidase mimicking activity of magnetic nanoparticles[J]. International Journal of Molecular Sciences, 2013,14(5):9999-10014.
doi: 10.3390/ijms14059999 URL pmid: 23665902 |
| [67] |
Cheng X, Huang L, Yang X, et al. Rational design of a stable peroxidase mimic for colorimetric detection of H2O2 and glucose:A synergistic CeO2/Zeolite Y nanocomposite[J]. Journal of Colloid and Interface Science, 2019,535:425-435.
doi: 10.1016/j.jcis.2018.09.101 URL pmid: 30317083 |
| [68] |
Li YZ, Li TT, Chen W, et al. Co4N nanowires:noble-metal-free peroxidase mimetic with excellent salt- and temperature-resistant abilities[J]. Acs Applied Materials & Interfaces, 2017,9(35):29881-29888.
doi: 10.1021/acsami.7b09861 URL pmid: 28806505 |
| [69] |
Su L, Feng J, Zhou X, et al. Colorimetric detection of urine glucose based ZnFe2O4 magnetic nanoparticles[J]. Analytical Chemistry, 2012,84(13):5753-5758.
doi: 10.1021/ac300939z URL pmid: 22702236 |
| [70] |
Jiang Y, Shi M, Liu Y, et al. Aptamer/AuNP biosensor for colorimetric profiling of exosomal proteins[J]. Angewandte Chemie-International Edition, 2017,56(39):11916-11920.
doi: 10.1002/anie.201703807 URL pmid: 28834063 |
| [71] |
Ma X, Song L, Zhou N, et al. A novel aptasensor for the colorimetric detection of S. typhimurium based on gold nanoparticles[J]. International Journal of Food Microbiology, 2017,245:1-5.
doi: 10.1016/j.ijfoodmicro.2016.12.024 URL pmid: 28107686 |
| [72] |
Kim YJ, Kim HS, Chon JW, et al. New colorimetric aptasensor for rapid on-site detection of Campylobacter jejuni and Campylobacter coli in chicken carcass samples[J]. Analytica Chimica Acta, 2018,1029:78-85.
doi: 10.1016/j.aca.2018.04.059 URL pmid: 29907294 |
| [73] |
Lee B, Park JH, Byun JY, et al. An optical fiber-based LSPR aptasensor for simple and rapid in-situ detection of ochratoxin A[J]. Biosensors & Bioelectronics, 2018,102:504-509.
doi: 10.1016/j.bios.2017.11.062 URL pmid: 29197812 |
| [74] |
Liu LH, Zhou XH, Shi HC. Portable optical aptasensor for rapid detection of mycotoxin with a reversible ligand-grafted biosensing surface[J]. Biosensors & Bioelectronics, 2015,72:300-305.
doi: 10.1016/j.bios.2015.05.033 URL pmid: 26000463 |
| [75] | Weisshoff H, Wenzel K, Schulze-Rothe S, et al. Characterization of aptamer bc 007 substance and product using circular dichroism and nuclear magnetic resonance spectroscopy[J]. Journal of Pharmaceutical Sciences, 2018,107(8):2033-2041. |
| [76] | Chavez JL, Hagen JA, Kelley-Loughnane N. Fast and selective plasmonic serotonin detection with aptamer-gold nanoparticle conjugates[J]. Sensors, 2017,17(4):681. |
| [77] |
Belal ASF, Ismail A, Elnaggar MM, et al. Click chemistry inspired copper sulphide nanoparticle-based fluorescence assay of kanamycin using DNA aptamer[J]. Spectrochimica Acta A Molecular and Biomolecular Spectroscopy, 2018,205:48-54.
doi: 10.1016/j.saa.2018.07.011 URL pmid: 30007899 |
| [78] |
Li W, Dong Y, Wang X, et al. PolyA-tailed and fluorophore-labeled aptamer-gold nanoparticle conjugate for fluorescence turn-on bioassay using iodide-induced ligand displacement[J]. Biosensors & Bioelectronics, 2015,66:43-49.
doi: 10.1016/j.bios.2014.10.047 URL pmid: 25460880 |
| [79] |
Wang X, Huang Y, Wu S, et al. Simultaneous detection of Staphylococcus aureus and Salmonella typhimurium using multicolor time-resolved fluorescence nanoparticles as labels[J]. International Journal of Food Microbiology, 2016,237:172-179.
doi: 10.1016/j.ijfoodmicro.2016.08.028 URL pmid: 27592261 |
| [80] |
Muniandy S, Dinshaw IJ, Teh SJ, et al. Graphene-based label-free electrochemical aptasensor for rapid and sensitive detection of foodborne pathogen[J]. Analytical and Bioanalytical Chemistry, 2017,409(29):6893-6905.
doi: 10.1007/s00216-017-0654-6 URL pmid: 29030671 |
| [81] |
Pang S, Labuza TP, He L. Development of a single aptamer-based surface enhanced Raman scattering method for rapid detection of multiple pesticides[J]. Analyst, 2014,139(8):1895-1901.
doi: 10.1039/c3an02263c URL pmid: 24551875 |
| [82] |
Duan N, Chang B, Zhang H, et al. Salmonella typhimurium detection using a surface-enhanced Raman scattering-based aptasensor[J]. International Journal of Food Microbiology, 2016,218:38-43.
URL pmid: 26599860 |
| [83] |
Wang Y, Wang Y, Li D, et al. Detection of nucleic acids and elimination of carryover contamination by using loop-mediated isothermal amplification and antarctic thermal sensitive uracil-DNA-glycosylase in a lateral flow biosensor:application to the detection of Streptococcus pneumoniae[J]. Microchimica Acta, 2018,185(4):212.
doi: 10.1007/s00604-018-2723-8 URL pmid: 29594577 |
| [84] |
Wu W, Yu L, Fang Z, et al. A lateral flow biosensor for the detection of human pluripotent stem cells[J]. Analytical Biochemistry, 2013,436(2):160-164.
doi: 10.1016/j.ab.2013.01.034 URL pmid: 23416186 |
| [85] |
Raston NH, Van-Thuan N, Gu MB. A new lateral flow strip assay(LFSA)using a pair of aptamers for the detection of Vaspin[J]. Biosensors & Bioelectronics, 2017,93:21-25.
doi: 10.1016/j.bios.2016.11.061 URL pmid: 27916536 |
| [1] | 陈彩萍, 任昊, 龙腾飞, 何冰, 鲁兆祥, 孙坚. 大肠杆菌Nissle 1917对炎症性肠病治疗作用的研究进展[J]. 生物技术通报, 2023, 39(6): 109-118. |
| [2] | 李典典, 粟元, 李洁, 许文涛, 朱龙佼. 抗菌适配体的筛选与应用进展[J]. 生物技术通报, 2023, 39(6): 126-132. |
| [3] | 李天顺, 李宸葳, 王佳, 朱龙佼, 许文涛. 功能核酸筛选过程中次级文库的有效制备[J]. 生物技术通报, 2023, 39(3): 116-122. |
| [4] | 唐瑞琪, 赵心清, 朱笃, 汪涯. 大肠杆菌对木质纤维素水解液抑制物的胁迫耐受性[J]. 生物技术通报, 2023, 39(11): 205-216. |
| [5] | 李仁瀚, 张乐乐, 刘春立, 刘秀霞, 白仲虎, 杨艳坤, 李业. 基于紫色杆菌素生物合成途径的L-色氨酸生物传感器的构建[J]. 生物技术通报, 2023, 39(10): 80-92. |
| [6] | 陈晓琳, 刘洋儿, 许文涛, 郭明璋, 刘慧琳. 合成生物学细胞传感技术在食品安全快速检测中的应用[J]. 生物技术通报, 2023, 39(1): 137-149. |
| [7] | 程深伟, 张克强, 梁军锋, 刘福元, 郜兴亮, 杜连柱. 畜禽养殖粪污中典型致病菌的三重微滴式数字PCR定量检测方法的建立[J]. 生物技术通报, 2022, 38(9): 271-280. |
| [8] | 刘理慧, 储锦华, 隋雨欣, 陈杨, 程古月. 沙门氏菌中主要毒力因子的研究进展[J]. 生物技术通报, 2022, 38(9): 72-83. |
| [9] | 高伟欣, 黄火清, 赵晶, 张鑫, 杨宁, 杨浩萌. 应用于基因编辑的核糖核蛋白复合体的构建与活性验证[J]. 生物技术通报, 2022, 38(8): 60-68. |
| [10] | 周子琦, 张洋子, 兰欣悦, 刘洋儿, 朱龙佼, 许文涛. 发光核酸适配体的筛选及应用[J]. 生物技术通报, 2022, 38(5): 240-247. |
| [11] | 王子琰, 王建, 张伦, 桂水清, 卢雪梅. 家蝇抗菌肽MDC对鼠伤寒沙门氏菌的抑菌稳定性研究[J]. 生物技术通报, 2022, 38(3): 149-156. |
| [12] | 孙曼銮, 葛赛, 卜佳, 朱壮彦. 大肠杆菌核糖核酸酶调控机制研究[J]. 生物技术通报, 2022, 38(3): 234-245. |
| [13] | 兰欣悦, 刘宁宁, 朱龙佼, 陈旭, 褚华硕, 李相阳, 段诺, 许文涛. 四环素双价适配体非酶免标记传感器[J]. 生物技术通报, 2022, 38(3): 276-284. |
| [14] | 张雅涵, 朱丽霞, 胡静, 朱亚静, 张雪婧, 曹叶中. 草甘膦在我国生物育种产业化应用中的机遇与挑战[J]. 生物技术通报, 2022, 38(11): 1-9. |
| [15] | 刘宁宁, 王鑫昕, 兰欣悦, 褚华硕, 陈旭, 常世敏, 李腾飞, 许文涛. G-三链体可视化核酸传感器用于四环素的检测[J]. 生物技术通报, 2022, 38(10): 106-114. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||