








生物技术通报 ›› 2021, Vol. 37 ›› Issue (8): 275-283.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1247
贺小丽1,3( ), 郭磊周1,2, 韩佳慧2, 唐殷1,2, 袁媛1,2, 代其林1, 平淑珍2, 江世杰1(
), 郭磊周1,2, 韩佳慧2, 唐殷1,2, 袁媛1,2, 代其林1, 平淑珍2, 江世杰1( )
)
收稿日期:2020-10-11
出版日期:2021-08-26
发布日期:2021-09-10
作者简介:贺小丽,女,硕士研究生,研究方向:特殊环境微生物功能基因资源利用;E-mail: 基金资助:
HE Xiao-li1,3( ), GUO Lei-zhou1,2, HAN Jia-hui2, TANG Yin1,2, YUAN Yuan1,2, DAI Qi-lin1, PING Shu-zhen2, JIANG Shi-jie1(
), GUO Lei-zhou1,2, HAN Jia-hui2, TANG Yin1,2, YUAN Yuan1,2, DAI Qi-lin1, PING Shu-zhen2, JIANG Shi-jie1( )
)
Received:2020-10-11
Published:2021-08-26
Online:2021-09-10
摘要:
细菌脂蛋白是一种脂质修饰的膜蛋白,参与细胞膜合成等多种重要生理过程。脂蛋白形成过程依赖于Lol转运系统,该蛋白最先在细胞质中以前体的形式合成,然后在细胞膜上被加工为成熟脂蛋白,锚定于细菌外膜周质侧。Lol系统由LolA-E五种蛋白组成,其中脂蛋白在周质空间中依赖伴侣蛋白 LolA进行转运,LolA将脂蛋白以“mouth to mouth”的方式从LolCDE转运至LolB,进而完成脂蛋白定位。重点对周质分子伴侣LolA结构、参与的转运体系及其生物学功能进行综述,旨在通过对脂蛋白转运分子机制的理解为感染性疾病的治疗提供更多的药物靶点。
贺小丽, 郭磊周, 韩佳慧, 唐殷, 袁媛, 代其林, 平淑珍, 江世杰. 细菌周质分子伴侣LolA研究进展[J]. 生物技术通报, 2021, 37(8): 275-283.
HE Xiao-li, GUO Lei-zhou, HAN Jia-hui, TANG Yin, YUAN Yuan, DAI Qi-lin, PING Shu-zhen, JIANG Shi-jie. Research Progress on Bacterial Periplasmic Chaperone LolA[J]. Biotechnology Bulletin, 2021, 37(8): 275-283.

图1 大肠杆菌LolA的晶体结构[22] LolA的疏水腔由开放的β-桶和α-螺旋盖子组成,也是脂质的结合位点;LolA的C-末端环由短α-螺旋和第12位β-链组成,能够正确地将脂蛋白递送至OM
Fig. 1 Crystal structure of Escherichia coli LolA[22] The hydrophobic cavity of LolA is composed of an open β-barrel and an α-helical lid,which is also a binding site for lipids;the C-terminal loop of LolA is composed of a short α-helix and a twelfth β-strand,which can correctly combine lipoprotein delivery to OM

图2 Lol系统介导的脂蛋白转运和外膜锚定[22] a:脂蛋白首先置于LolE中(1),然后转移至位于LolC上的LolA(2)。当LolCDE结合脂蛋白时,LolA和LolCDE之间的相互作用增加(实线箭头)。然后LolA以ATP依赖性方式与脂蛋白形成亲水性复合物,同时打开LolA疏水腔(3)。b:在穿过周质空间后,LolA和LolB以“mouth to mouth”的方式互作将脂蛋白从LolA转移至LolB(4),最终定位到外膜
Fig. 2 Lipoprotein transport and outer membrane anchoring mediated by the Lol system[22] a:Lipoprotein is first placed in LolE(1),and then transferred to LolA located on LolC(2). When LolCDE binds to lipoproteins,the interaction between LolA and LolCDE increases(solid arrow). Then LolA forms a hydrophilic complex with lipoproteins in an ATP-dependent manner,and opens the hydrophobic cavity of LolA at the same time(3). b:After passing through the periplasmic space,LolA and LolB interact in a “mouth to mouth” manner to transfer lipoproteins from LolA to LolB(4),and finally locate to the outer membrane
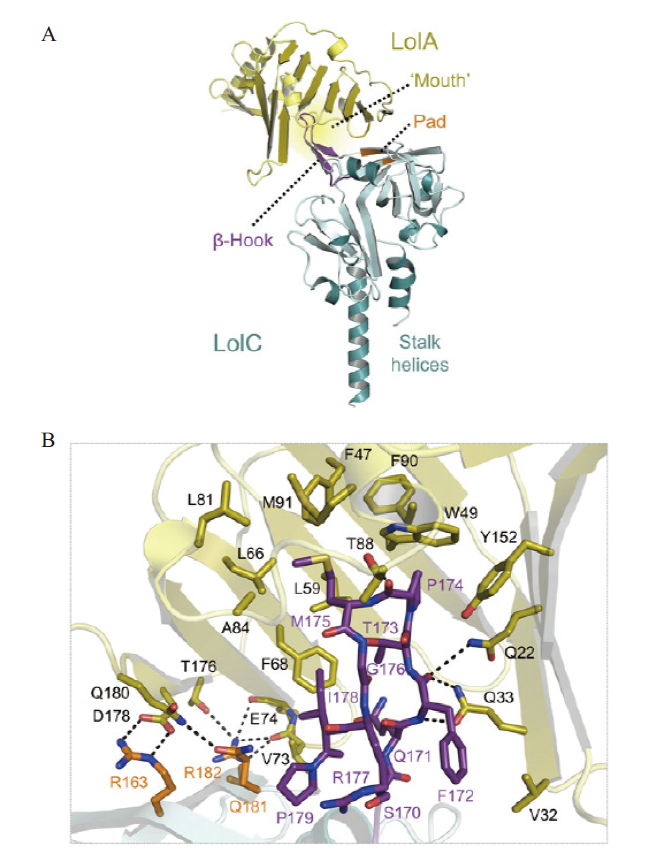
图3 LolA的晶体结构与LolC的周质域结合[30] A:LolA - LolC 复合体的总体结构;B:互作界面视图。LolC 和 LolA 分别以青色和金色表示。属于 Hook 和 Pad 的残留物显示为紫色和橙色。与 LolC交互的内容以棒状显示
Fig. 3 Crystal structure of LolA is combined with the periplasmic domain of LolC[30] A:The overall structure of the LolA-LolC complex. B:Interaction interface view. LolC and LolA are represented in cyan and gold,respectively. The residue belonging to Hook and Pad is shown in purple and orange. The content that interacts with LolC is displayed in a bar shape
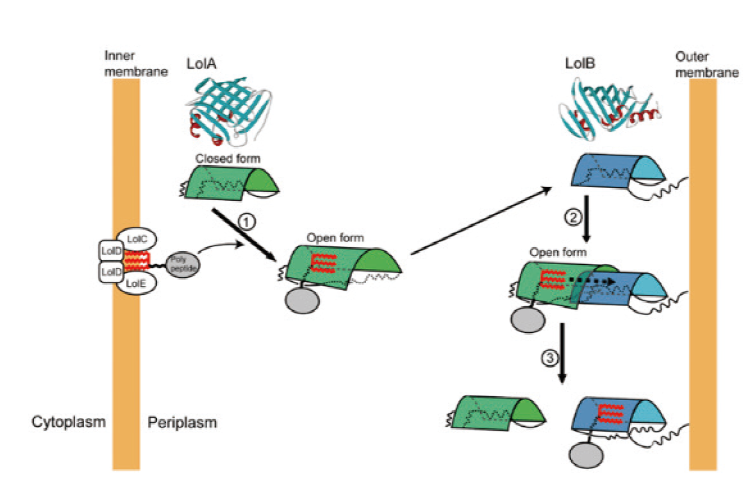
图4 脂蛋白在LolA和LolB上的转运模型[36] LolA 和 LolB 分别用绿色和蓝色的桶状表示,红色代表酰基链。第 1 步,从内膜周质侧的 LolCDE 获得一种脂蛋白,LolA 经过构象变化并使其疏水腔适应了 1-3 种脂蛋白酰基链。第 2 步,脂蛋白-LolA 复合物与锚定在外膜上的LolB 相互作用,在该复合物中,LolA 和 LolB 形成一个疏水的、类似通道的结构。第 3 步,在脂蛋白酰基链通过这两个蛋白形成的疏水通道从 LolA 转移到 LolB
Fig. 4 Transport model of lipoprotein on LolA and LolB[36] LolA and LolB are represented by green and blue barrels,respectively,and red represents the acyl chain. Step 1: a lipoprotein is obtained from LolCDE on the periplasmic side of the inner membrane. LolA undergoes a conformational change and adapts its hydrophobic cavity to 1 to 3 lipoprotein acyl chains. Step 2: the lipoprotein-LolA complex interacts with the LolB anchored on the outer membrane. In the complex,LolA and LolB form a hydrophobic,channel-like structure. Step 3: the acyl chain of the lipoprotein is transferred from LolA to LolB through the hydrophobic channel formed by these two proteins

图5 LolA疏水腔中BLP的结合模式[47] A:左:启动配置,其中 BLP(黄色)位于 LolA(青色)腔的外部。右上:BLP 的3个脂尾在腔口附近结合的结合方式。右下:一个脂尾在腔深,而另两个在腔口处;B:3个独立模拟的 BLP 脂尾和 F90 残基之间的最小距离(如插图中范德瓦尔斯表示法所示)
Fig.5 Binding mode of BLP in the hydrophobic cavity of LolA[47] A:Left:Start configuration,where BLP(yellow)is located outside the LolA(cyan)cavity. Upper right:The combination of the three fat tails of BLP near the mouth of the cavity. Lower right:One fat tail is deep in the cavity,and the other two are at the mouth of the cavity. B:The minimum distance between the three independently simulated BLP fat tails and F90 residues(as shown in the illustration by Van der Waals notation)
| [1] |
Sankaran K, Wu HC. Lipid modification of bacterial Prolipoprotei. Transfer of diacylglyceryl moiety from phosphatidylglycerol[J]. Journal of Biological Chemistry, 1994, 269(31):19701-19706.
pmid: 8051048 |
| [2] |
Buddelmeijer N. The molecular mechanism of bacterial lipoprotein modification--how, when and why?[J]. FEMS Microbiol Rev 2015, 39(2):246-261.
doi: 10.1093/femsre/fuu006 pmid: 25670733 |
| [3] |
Lovullo ED, Wright LF, Isabella V, et al. Revisiting the Gram-negative lipoprotein paradigm[J]. J Bacteriol, 2015, 197(10):1705-1715.
doi: 10.1128/JB.02414-14 URL |
| [4] |
Hutchings MI, Palmer T, Harrington DJ, et al. Lipoprotein biogenesis in Gram-positive bacteria:knowing when to hold'em, knowing when to fold 'em[J]. Trends Microbiol, 2009, 17(1):13-21.
doi: 10.1016/j.tim.2008.10.001 pmid: 19059780 |
| [5] |
Nakayama H, Kurokawa K, Lee BL. Lipoproteins in bacteria:structures and biosynthetic pathways[J]. FEBS J, 2012, 279(23):4247-4268.
doi: 10.1111/febs.12041 pmid: 23094979 |
| [6] |
Schenk M, Belisle JT, Modlin RL. TLR2 looks at lipoproteins[J]. Immunity, 2009, 31(6):847-849.
doi: 10.1016/j.immuni.2009.11.008 URL |
| [7] |
Bernadac A, Gavioli M, Lazzaroni JC, et al. Escherichia coli tol-pal mutants form outer membrane vesicles[J]. J Bacteriol, 1998, 180:4872-4878.
pmid: 9733690 |
| [8] |
Clavel T, Germon P, Vianney A, et al. Tol B protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA[J]. Mol Microbiol, 1998, 29:359-367.
pmid: 9701827 |
| [9] |
Ehrmann M, Ehrle R, Hofmann E, et al. The ABC maltose transporter[J]. Mol Microbiol, 1998, 29:685-694.
pmid: 9723909 |
| [10] |
Nikaido H. Multiple antibiotic resistance and efflux[J]. Curr Opin Microbiol, 1998, 1:516-523.
pmid: 10066525 |
| [11] |
Tokuda H. Biogenesis of outer membranes in Gram-negative bacteria[J]. Bioscience, Biotechnology, and Biochemistry, 2009, 73(3):465-473.
doi: 10.1271/bbb.80778 URL |
| [12] |
Doerrler WT. Lipid trafficking to the outer membrane of Gram-negative bacteria[J]. Molecular Microbiology, 2006, 60(3):542-552.
pmid: 16629659 |
| [13] | Lorenz C, Dougherty TJ, Lory S. Correct sorting of lipoproteins into the inner and outer membranes of Pseudomonas aeruginosa by the Escherichia coli LolCDE transport system[J]. mBio, 2019, 10(2):e00194-19. |
| [14] | Narita S, Tokuda H. Bacterial lipoproteins;biogenesis, sorting and quality control[J]. Biochimica et Biophysica Acta(BBA)-Molec-ular and Cell Biology of Lipids, 2017, 1862(11):1414-1423. |
| [15] | Zückert WR. Secretion of bacterial lipoproteins:through the cytoplasmic membrane, the periplasm and beyond[J]. Biochimica et Biophysica Acta(BBA)-Molecular Cell Research, 2014, 1843(8):1509-1516. |
| [16] |
Matsuyama S, Tajima T, Tokuda H. A novel periplasmic carrier protein involved in the sorting and transport of Escherichia coli lipoproteins destined for the outer membrane[J]. The EMBO Journal, 1995, 14(14):3365.
doi: 10.1002/embj.1995.14.issue-14 URL |
| [17] |
Konovalova A, Silhavy TJ. Outer membrane lipoprotein biogenesis:Lol is not the end[J]. Philos Trans R Soc Lond B Biol Sci, 2015, 370:20150030.
doi: 10.1098/rstb.2015.0030 URL |
| [18] | Goemans C, Denoncin K, Collet JF. Folding mechanisms of periplasmic proteins[J]. Biochimica et Biophysica Acta(BBA)-Molecular Cell Research, 2014, 1843(8):1517-1528. |
| [19] |
Takeda K, Miyatake H, Yokota N, et al. Crystal structures of bacterial lipoprotein localization factors, LolA and LolB[J]. EMBO J, 2003, 22:3199-3209.
doi: 10.1093/emboj/cdg324 URL |
| [20] |
Okuda S, Watanabe S, Tokuda H. A short helix in the C-terminal region of LolA is important for the specific membrane localization of lipoproteins[J]. FEBS Lett, 2008, 582:2247-2251.
doi: 10.1016/j.febslet.2008.05.022 URL |
| [21] |
Murahari P, Anishetty S, Pennathur G, et al. Research article:Understanding the lid movements of LolA in Escherichia coli using molecular dynamics simulation and in silico point mutation[J]. Computational Biology and Chemistry, 2013, 47:71-80.
doi: 10.1016/j.compbiolchem.2013.06.005 URL |
| [22] |
Okuda S, Tokuda H. Lipoprotein Sorting in Bacteria[J]. Annu Rev Microbiol, 2011, 65(1):239-259.
doi: 10.1146/annurev-micro-090110-102859 URL |
| [23] |
Pastukhov AV, Ropson IJ. Fluorescent dyes as probes to study lipid-binding proteins[J]. Proteins, 2003, 53:607-615.
doi: 10.1002/(ISSN)1097-0134 URL |
| [24] |
Oguchi Y, Takeda K, Watanabe S, et al. Opening and closing of the hydrophobic cavity of LolA coupled to lipoprotein binding and release[J]. J Biol Chem, 2008, 283:25414-25420.
doi: 10.1074/jbc.M804736200 pmid: 18617521 |
| [25] |
Tao K, Watanabe S, Narita SI, et al. A periplasmic LolA derivative with a lethal disulfide bond activates the Cpx stress response system[J]. Journal of Bacteriology, 2010, 192(21):5657-5662.
doi: 10.1128/JB.00821-10 URL |
| [26] |
Okuda S, Tokuda H. Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB[J]. Proceedings of the National Academy of Sciences, 2009, 106(14):5877-5882.
doi: 10.1073/pnas.0900896106 URL |
| [27] |
Mizutani M, Mukaiyama K, Xiao J, et al. Functional differentiation of structurally similar membrane subunits of the ABC transporter LolCDE complex[J]. FEBS letters, 2013, 587(1):23-29.
doi: 10.1016/j.febslet.2012.11.009 URL |
| [28] |
Narita S, Tanaka K, Matsuyama S, et al. Disruption of lolCDE, encoding an ATP-binding cassette transporter, is lethal for Escherichia coli and prevents release of lipoproteins from the inner membrane[J]. Journal of Bacteriology, 2002, 184(5):1417-1422.
doi: 10.1128/JB.184.5.1417-1422.2002 URL |
| [29] |
Yasuda M, Iguchi-Yokoyama A, Matsuyama SI, et al. Membrane topology and functional importance of the periplasmic region of ABC transporter LolCDE[J]. Bioscience, Biotechnology, and Biochemistry, 2009, 73(10):2310-2316.
doi: 10.1271/bbb.90451 URL |
| [30] | Kaplan E, Greene NP, Crow A, et al. Insights into bacterial lipoprotein trafficking from a structure of LolA bound to the LolC periplasmic domain[J]. Proceedings of the National Academy of Sciences of the United States of America, 2018, 115(31):E7389-E7397. |
| [31] |
Watanabe S, Oguchi Y, Yokota N, et al. Large-scale preparation of the homogeneous LolA-lipoprotein complex and efficient in vitro transfer of lipoproteins to the outer membrane in a LolB-dependent manner[J]. Protein Science, 2007, 16(12):2741-2749.
doi: 10.1110/(ISSN)1469-896X URL pmid: 18029423 |
| [32] |
Ito Y, Kanamaru K, Taniguchi N, et al. A novel ligand bound ABC transporter, LolCDE, provides insights into the molecular mechanisms underlying membrane detachment of bacterial lipoproteins[J]. Molecular Microbiology, 2006, 62(4):1064-1075.
doi: 10.1111/mmi.2006.62.issue-4 URL |
| [33] |
Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli[J]. Nature methods, 2006, 3(4):263-265.
doi: 10.1038/nmeth864 URL |
| [34] |
Wang L, Xie J, Schultz PG. Expanding the genetic code[J]. Annu Rev Biophys Biomol Struct, 2006, 35:225-249.
doi: 10.1146/annurev.biophys.35.101105.121507 URL |
| [35] |
Szewczyk J, Collet JF. The journey of lipoproteins through the cell:one birthplace, multiple destinations[J]. Adv Microb Physiol, 2016, 69:1-50.
doi: S0065-2911(16)30024-8 pmid: 27720009 |
| [36] |
Nakada S, Sakakura M, Takahashi H, et al. Structural investigation of the interaction between LolA and LolB using NMR[J]. Journal of Biological Chemistry, 2009, 284(36):24634-24643.
doi: 10.1074/jbc.M109.001149 URL |
| [37] |
Matsuyama SI, Yokota N, Tokuda H. A novel outer membrane lipoprotein, LolB(Hem M), involved in the LolA(p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli[J]. EMBO J, 1997, 16:6947-6955.
pmid: 9384574 |
| [38] |
Yokota N, Kuroda T, Matsuyama S, et al. Characterization of the Lol A-Lol B system as the general lipoprotein localization mechanism of Escherichia coli[J]. J Biol Chem, 1999, 274(43):30995-30999.
pmid: 10521496 |
| [39] |
Masuda K, Matsuyama S, Tokuda H. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization[J]. Proc Natl Acad Sci USA, 2002, 99(11):7390-7395.
doi: 10.1073/pnas.112085599 URL |
| [40] |
Tajima T, Yokota N, Matsuyama S, et al. Genetic analyses of the in vivo function of LolA, a periplasmic chaperone involved in the outer membrane localization of Escherichia coli lipoproteins[J]. FEBS Letters, 1998, 439(1-2):51-54.
pmid: 9849875 |
| [41] |
Miyamoto A, Matsuyama S, Tokuda H. Mutant of LolA, a lipoprotein-specific molecular chaperone of Escherichia coli, defective in the transfer of lipoproteins to LolB[J]. Biochem Biophys Res Commun, 2001, 287(5):1125-1128.
doi: 10.1006/bbrc.2001.5705 URL |
| [42] |
Tao K, Narita S, Tokuda H, et al. Defective lipoprotein sorting induces lolA expression through the rcs stress response phosphorelay system[J]. Journal of Bacteriology, 2012, 194(14):3643-3650.
doi: 10.1128/JB.00553-12 URL |
| [43] |
Miyamoto A, Matsuyama SI, Tokuda H. Dominant negative mutant of a lipoprotein-specific molecular chaperone, LolA, tightly associates with LolCDE[J]. FEBS Letters, 2002, 528(1/3):193-196.
doi: 10.1016/S0014-5793(02)03305-7 URL |
| [44] |
Grabowicz M, Silhavy TJ. Redefining the essential trafficking pathway for outer membrane lipoproteins[J]. Proc Natl Acad Sci USA, 2017, 114(18):4769-4774.
doi: 10.1073/pnas.1702248114 URL |
| [45] |
Choi U, Lee C. Antimicrobial agents that inhibit the outer membrane assembly machines of gram-negative bacteria[J]. Journal of Microbiology and Biotechnology, 2019, 29(1):1-10.
doi: 10.4014/jmb.1804.03051 URL |
| [46] |
Muheim C, Gotzke H, Eriksson A, et al. Increasing the permeability of Escherichia coli using MAC13243[J]. Scientific Reports, 2017, 7(1):17629.
doi: 10.1038/s41598-017-17772-6 URL |
| [47] |
Boags AT, Samsudin F, Khalid S, et al. Details of hydrophobic entanglement between small molecules and Braun’s lipoprotein within the cavity of the bacterial chaperone LolA[J]. Scientific Reports, 2019, 9(1):3717.
doi: 10.1038/s41598-019-40170-z URL |
| [48] |
Bosch V, Braun V. Distribution of murein-lipoprotein between the cytoplasmic and outer membrane of Escherichia coli[J]. FEBS Lett, 1973, 34(2):307-310.
pmid: 4583850 |
| [49] |
Shu W, Liu J, Ji H, et al. Core structure of the outer membrane lipoprotein from Escherichia coli at 1. 9 A resolution[J]. J Mol Biol, 2000, 299(4):1101-1112.
pmid: 10843861 |
| [50] |
Watanabe S, Oguchi Y, Takeda K, et al. Introduction of a lethal redox switch that controls the opening and closing of the hydrophobic cavity in LolA[J]. Journal of Biological Chemistry, 2008, 283(37):25421-25427.
doi: 10.1074/jbc.M804737200 pmid: 18621730 |
| [51] |
Pathania R, Zlitni S, Barker C, et al. Chemical genomics in Escherichia coli identifies an inhibitor of bacterial lipoprotein targeting[J]. Nat Chem Biol, 2009, 5(11):849-856.
doi: 10.1038/nchembio.221 pmid: 19783991 |
| [52] |
Narita S, Tanaka K, Matsuyama S, et al. Disruption of lolCDE encoding an ATP-binding-cassette transporter is lethal for Escherichia coli and prevents the release of lipoproteins from the inner membrane[J]. J Bacteriol, 2002, 184(5):1417-1422.
doi: 10.1128/JB.184.5.1417-1422.2002 URL |
| [53] |
Juncker AS, Willenbrock H, Von Heijne G, et al. Prediction of lipoprotein signal peptides in gram-negative bacteria[J]. Protein Sci, 2003, 12(8):1652-1662.
doi: 10.1110/ps.0303703 URL |
| [54] |
Lewenza S, Vidal-Ingigliardi D, Pugsley AP. Direct visualization of red fluorescent lipoproteins indicates conservation of the membrane sorting rules in the family Enterobacteriaceae[J]. J Bacteriol, 2006, 188(10):3516-3524.
pmid: 16672606 |
| [55] | Grabowicz M. Lipoproteins and their trafficking to the outer membrane[J]. EcoSal Plus, 2019, 8:0038. |
| [56] |
Schulze R J, Chen SY, Kumru O S, et al. Translocation of Borrelia burgdorferi surface lipoprotein OspA through the outer membrane requires an unfolded conformation and can initiate at the C-terminus[J]. Mol Microbiol, 2010, 76(5):1266-1278.
doi: 10.1111/j.1365-2958.2010.07172.x pmid: 20398211 |
| [57] |
Rhodes RG, Samarasam MN, Van Groll EJ, et al. Mutations in Flavobacterium johnsoniae sprE result in defects in gliding motility and protein secretion[J]. J Bacteriol, 2011, 193(19):5322-5327.
doi: 10.1128/JB.05480-11 pmid: 21784937 |
| [58] |
Liao CT, Chiang YC, Hsiao YM. Functional characterization and proteomic analysis of lolA in Xanthomonas campestris pv. Campestris[J]. BMC Microbiology, 2019, 19(1):20.
doi: 10.1186/s12866-019-1387-9 URL |
| [1] | 董海娇, 杨晓玉, 莫蓓莘, 陈雪梅, 崔洁. 核糖核酸5'端NAD+帽子修饰研究进展[J]. 生物技术通报, 2022, 38(2): 245-251. |
| [2] | 赵杰, 李安, 梁刚, 靳欣欣, 潘立刚. 植物环状RNA的研究新进展[J]. 生物技术通报, 2022, 38(10): 1-9. |
| [3] | 王志山, 黎妮, 王伟平, 刘洋. 水稻种子内生细菌研究进展[J]. 生物技术通报, 2022, 38(1): 236-246. |
| [4] | 薛翔澜, 丁洋洋, 刘悦, 李晓波, 蒋琳, 何晓红, 马月辉, 赵倩君. 哺乳动物m6A与生长发育相关生物学功能研究进展[J]. 生物技术通报, 2021, 37(4): 251-259. |
| [5] | 冯逸龙, 张文利. DNA鸟嘌呤四联体研究进展[J]. 生物技术通报, 2020, 36(7): 23-31. |
| [6] | 黄幸, 丁峰, 彭宏祥, 潘介春, 何新华, 徐炯志, 李琳. 植物WRKY转录因子家族研究进展[J]. 生物技术通报, 2019, 35(12): 129-143. |
| [7] | 王春雨, 张茜. 植物NAC转录因子功能研究进展[J]. 生物技术通报, 2018, 34(11): 8-14. |
| [8] | 徐海冬, 冷奇颖, PATRICIAAdu-Asiamah, 王章, 李婷, 张丽. 环状RNA的特征及其在畜禽中的研究进展[J]. 生物技术通报, 2018, 34(11): 56-69. |
| [9] | 包琦, 赵凯, 段子渊. 猪Btnl5基因的克隆、表达分析及对NF-κB信号通路的调控作用[J]. 生物技术通报, 2017, 33(6): 142-148. |
| [10] | 钟瑞春,李婷婷,唐荣华,王兴军,李翠,侯蕾,赵传志. 花生温度诱导载脂蛋白基因AhTIL1的克隆和表达研究[J]. 生物技术通报, 2016, 32(4): 102-109. |
| [11] | 罗燕,刘小刚,周志钦. 植物糖基转移酶基因的分离方法及其生物学功能研究进展[J]. 生物技术通报, 2016, 32(12): 34-39. |
| [12] | 牛旭龙, 冯万军, 马金虎, 邢国芳. 植物长链非编码RNA功能研究进展[J]. 生物技术通报, 2015, 31(6): 1-7. |
| [13] | 姜珊珊, 张丹, 孔祥培, 周严, 李德全. 植物中的钙依赖蛋白激酶(CDPK)的结构特征和功能研究进展[J]. 生物技术通报, 2013, 0(6): 12-19. |
| [14] | 谭亚清, 刘德虎. 人酸性成纤维细胞生长因子的研究进展[J]. 生物技术通报, 2013, 0(5): 22-27. |
| [15] | 倪博, 白方方, 刘茂军, 冯志新, 熊祺琰, 魏建忠, 邵国青. 支原体脂蛋白及其变异与宿主互作研究进展[J]. 生物技术通报, 2013, 0(2): 49-54. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||