








生物技术通报 ›› 2021, Vol. 37 ›› Issue (4): 251-259.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1358
薛翔澜1( ), 丁洋洋1, 刘悦1, 李晓波2, 蒋琳1, 何晓红1, 马月辉1, 赵倩君1(
), 丁洋洋1, 刘悦1, 李晓波2, 蒋琳1, 何晓红1, 马月辉1, 赵倩君1( )
)
收稿日期:2020-11-06
出版日期:2021-04-26
发布日期:2021-05-13
作者简介:薛翔澜,女,硕士研究生,研究方向:动物遗传育种;E-mail:基金资助:
XUE Xiang-lan1( ), DING Yang-yang1, LIU Yue1, LI Xiao-bo2, JIANG Lin1, HE Xiao-hong1, MA Yue-hui1, ZHAO Qian-jun1(
), DING Yang-yang1, LIU Yue1, LI Xiao-bo2, JIANG Lin1, HE Xiao-hong1, MA Yue-hui1, ZHAO Qian-jun1( )
)
Received:2020-11-06
Published:2021-04-26
Online:2021-05-13
摘要:
RNA 表观遗传修饰N6-甲基腺嘌呤(N6-methyladenosine,m6A)是RNA中存在最为广泛的中间化学修饰,普遍存在于生命体中,其中以哺乳动物中的研究最为广泛。m6A修饰酶和结合蛋白的发现证明了RNA甲基化修饰是动态可逆过程,常见的m6A修饰酶主要包括甲基转移酶和去甲基化酶。近年研究陆续发现通过调控m6A在RNA修饰水平上影响mRNA剪切、翻译、稳定性、出核等过程。RNA甲基化介导的表观转录组学参与诸多生物学过程。结合m6A前沿研究成果,对m6A甲基化主要的修饰酶、检测方法及其在胚胎发育、肌肉发育与脂肪生成等方面的生物学功能进行综述,旨为探究m6A甲基化修饰在动物重要的生物学过程中的调控机制寻找新方向。
薛翔澜, 丁洋洋, 刘悦, 李晓波, 蒋琳, 何晓红, 马月辉, 赵倩君. 哺乳动物m6A与生长发育相关生物学功能研究进展[J]. 生物技术通报, 2021, 37(4): 251-259.
XUE Xiang-lan, DING Yang-yang, LIU Yue, LI Xiao-bo, JIANG Lin, HE Xiao-hong, MA Yue-hui, ZHAO Qian-jun. Research Progress on Biological Function Growth and Development Related to N6-methyladenosine in Mammals[J]. Biotechnology Bulletin, 2021, 37(4): 251-259.
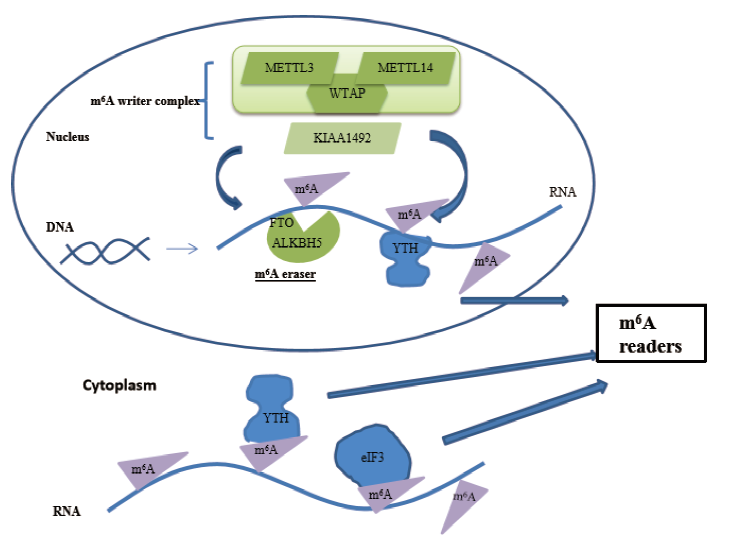
图2 m6A甲基化的调节过程 METTL3 -METTL14 -WTAP 复合物、KIAA1492 为Writers;FTO、ALKBH5为Erasers;YTH 结构域家族蛋白和eIF3 为Readers
Fig.2 The regulation process of the m6A METTL3-METTL14-WTAP complex and KIAA1492 are Writers. FTO, ALKBH5 are Erasers. YTH domain family proteins and eIF3 are Readers
| 检测方法 Detection method | 优点 Advantage | 缺点 Defect | 参考文献 Reference |
|---|---|---|---|
| 比色法 Colorimetry | RNA整体水平检测、成本低、操作简单 | 不具备特异性、低通量 | [32] |
| MeRIP-Seq | 高通量、单碱基、高灵敏度、量化转录片段 | 依赖抗体、无法定量、无法在全转录组水平精确定位 | [33-34] |
| HPLC-LC/MS/MS | 快速、灵敏、分辨率高、可准确定量定性分析 | 定性能力较弱、检测灵敏度一般 | [35] |
| m6A-miCLIP | 高通量、单碱基、UV诱导抗体-RNA交联 | 依赖于抗体 | [36] |
| PA-m6A-Seq | 高通量、单碱基、细胞水平定位 | 依赖于抗体 | [46] |
| m6A-LAIC-Seq | 高通量、精确定量 | 依赖于抗体 | [36] |
| m6A-REF-Seq/ MAZTER-Seq | 不依赖于抗体、特异性区分m6A修饰的内切核酸酶、高通量 | 不能覆盖所有m6A位点 | [44-45] |
表1 m6A甲基化检测手段
Table 1 Detection method of m6A methylation
| 检测方法 Detection method | 优点 Advantage | 缺点 Defect | 参考文献 Reference |
|---|---|---|---|
| 比色法 Colorimetry | RNA整体水平检测、成本低、操作简单 | 不具备特异性、低通量 | [32] |
| MeRIP-Seq | 高通量、单碱基、高灵敏度、量化转录片段 | 依赖抗体、无法定量、无法在全转录组水平精确定位 | [33-34] |
| HPLC-LC/MS/MS | 快速、灵敏、分辨率高、可准确定量定性分析 | 定性能力较弱、检测灵敏度一般 | [35] |
| m6A-miCLIP | 高通量、单碱基、UV诱导抗体-RNA交联 | 依赖于抗体 | [36] |
| PA-m6A-Seq | 高通量、单碱基、细胞水平定位 | 依赖于抗体 | [46] |
| m6A-LAIC-Seq | 高通量、精确定量 | 依赖于抗体 | [36] |
| m6A-REF-Seq/ MAZTER-Seq | 不依赖于抗体、特异性区分m6A修饰的内切核酸酶、高通量 | 不能覆盖所有m6A位点 | [44-45] |
| [1] |
Yoon KJ, Ringeling FR, Vissers C, et al. Temporal control of mammalian cortical neurogenesis by m6A methylation[J]. Cell, 2017, 171(4):877-889.e17.
doi: 10.1016/j.cell.2017.09.003 URL |
| [2] |
Shi H, Zhang X, Weng YL, et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1[J]. Nature, 2018,563(7730):249-253.
doi: 10.1038/s41586-018-0666-1 URL |
| [3] |
Zhu B, Gong Y, Shen L, et al. Total Panax notoginseng saponin inhibits vascular smooth muscle cell proliferation and migration and intimal hyperplasia by regulating WTAP/p16 signals via m6A modulation[J]. Biomed Pharmacother, 2020,124:109935.
doi: 10.1016/j.biopha.2020.109935 URL |
| [4] |
Tanaka T, Weisblum B. Systematic difference in the methylation of ribosomal ribonucleic acid from gram-positive and gram-negative bacteria[J]. J Bacteriol, 1975,123(2):771-774.
pmid: 807565 |
| [5] |
Munns TW, Sims HF, Liszewski MK. Immunospecific retention of oligonucleotides possessing N6-methyladenosine and 7-methylguanosine[J]. J Biol Chem, 1977,252(9):3102-3104.
pmid: 323262 |
| [6] |
Epstein P, Reddy R, Henning D, et al. The nucleotide sequence of nuclear U6(4. 7 S)RNA[J]. J Biol Chem, 1980,255(18):8901-8906.
pmid: 6773955 |
| [7] |
Donald TD, Robert HT. The methylation state of poly A-containing- messenger RNA from cultured hamster cells[J]. Nucleic Acids Research, 1975,2(10):1653-1668.
doi: 10.1093/nar/2.10.1653 URL |
| [8] |
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq[J]. Nature, 2012,485(7397):201-206.
doi: 10.1038/nature11112 pmid: 22575960 |
| [9] |
Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons[J]. Cell, 2012,149(7):1635-1646.
doi: 10.1016/j.cell.2012.05.003 pmid: 22608085 |
| [10] |
Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation[J]. Trends in Genetics, 2013,29(2):108-115.
doi: 10.1016/j.tig.2012.11.003 URL |
| [11] |
Alarcón C R, Goodarzi H, Lee H, et al. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events[J]. Cell, 2015,162(6):1299-1308.
doi: 10.1016/j.cell.2015.08.011 URL |
| [12] |
Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation[J]. Nature Chemical Biology, 2014,10(2):93-95.
doi: 10.1038/nchembio.1432 URL |
| [13] | Dai D, Wang H, Zhu L, et al. N6-methyladenosine links RNA metabolism to cancer progression[J]. Cell Death & Disease, 2018,9(2):124. |
| [14] |
Bokar JA, Shambaugh ME, Polayes D, et al. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA(N6-adenosine)-methyltransferase[J]. RNA, 1997,3(11):1233-1247.
pmid: 9409616 |
| [15] |
Zhang C, Chen Y, Sun B, et al. m6A modulates haematopoietic stem and progenitor cell specification[J]. Nature, 2017,549(7671):273-276.
doi: 10.1038/nature23883 URL |
| [16] |
Wang Y, Li Y, Toth JI, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells[J]. Nature Cell Biology, 2014,16(2):191-198.
doi: 10.1038/ncb2902 URL |
| [17] |
Ping XL, Sun BF, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase[J]. Cell Research, 2014,24(2):177-189.
doi: 10.1038/cr.2014.3 URL |
| [18] |
Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases[J]. Mol Cell, 2016,63(2):306-317.
doi: 10.1016/j.molcel.2016.05.041 URL |
| [19] |
Schwartz S, Mumbach M, Jovanovic M, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites[J]. Cell Reports, 2014,8(1):284-296.
doi: 10.1016/j.celrep.2014.05.048 pmid: 24981863 |
| [20] |
Huang Y, Yan J, Li Q, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5[J]. Nucleic Acids Res, 2015,43(1):373-384.
doi: 10.1093/nar/gku1276 URL |
| [21] |
Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity[J]. Nat Genet, 2007,39(6):724-726.
doi: 10.1038/ng2048 URL |
| [22] |
Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity[J]. Science, 2007,316(5826):889-894.
doi: 10.1126/science.1141634 URL |
| [23] |
Scuteri A, Sanna S, Chen WM, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits[J]. PLoS Genet, 2007,3(7):e115.
doi: 10.1371/journal.pgen.0030115 URL |
| [24] |
Jia GF, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO[J]. Nature Chemical Biology, 2011,7(12):885-887.
doi: 10.1038/nchembio.687 URL |
| [25] |
Alarcón CR, Hyeseung L, Hani G, et al. N6-methyladenosine marks primary microRNAs for processing[J]. Nature, 2015,519(7544):482-485.
doi: 10.1038/nature14281 URL |
| [26] |
Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability[J]. Nature, 2014,505(7481):117-120.
doi: 10.1038/nature12730 URL |
| [27] | Liu S, Li G, Li Q, et al. The roles and mechanisms of YTH domain-containing proteins in cancer development and progression[J]. American Journal of Cancer Research, 2020,10(4):1068-1084. |
| [28] |
Liu N, Dai Q, Zheng GQ, et al. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions[J]. Nature, 2015,518(7540):560-564.
doi: 10.1038/nature14234 URL |
| [29] |
Meyer KD, Patil DP, Zhou J, et al. 5' UTR m6A promotes Cap-independent translation[J]. Cell, 2015,163(4):999-1010.
doi: 10.1016/j.cell.2015.10.012 pmid: 26593424 |
| [30] |
Li A, Chen YS, Ping XL, et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation[J]. Cell Res, 2017,27(3):444-447.
doi: 10.1038/cr.2017.10 URL |
| [31] |
Kasowitz SD, Ma J, Anderson SJ, et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development[J]. PLoS Genetics, 2018,14(5):e1007412.
doi: 10.1371/journal.pgen.1007412 URL |
| [32] |
Lee M, Kim B, Kim VN. Emerging roles of RNA modification:m6A and U-tail[J]. Cell, 2014,158(5):980-987.
doi: 10.1016/j.cell.2014.08.005 URL |
| [33] |
Wang Y, Li Y, Yue M, et al. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications[J]. Nature Neuroscience, 2018,21(2):195-206.
doi: 10.1038/s41593-017-0057-1 URL |
| [34] |
Fu Y, Dominissini D, Rechavi G, et al. Gene expression regulation mediated through reversible m6A RNA methylation[J]. Nature Reviews Genetics, 2014,15(5):293-306.
doi: 10.1038/nrg3724 URL |
| [35] |
da Silva DVT, Baião DDS, de Oliveira Silva F, et al. Betanin, a natural food additive:stability, bioavailability, antioxidant and preservative ability assessments[J]. Molecules, 2019,24(3):458.
doi: 10.3390/molecules24030458 URL |
| [36] |
Arivazhagan A, Arora A, Hegde AS, et al. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance[J]. Oncogene, 2018,37(4):522-533.
doi: 10.1038/onc.2017.351 URL |
| [37] |
Wei CM, Moss B. Methylation of newly synthesized viral messenger RNA by an enzyme in vaccinia virus[J]. National Academy of Sciences, 1974,71(8):3014-3018.
doi: 10.1073/pnas.71.8.3014 URL |
| [38] | Rottman FM, Desrosiers RC, Friderici K. Nucleotide methylation patterns in eukaryotic mRNA[J]. Chinese Bulletin of Life Sciences, 1976,19:539-550. |
| [39] |
Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from novikoff hepatoma cells[J]. National Academy of Sciences, 1974,71(10):3971-3975.
doi: 10.1073/pnas.71.10.3971 URL |
| [40] |
Salditt-Georgieff M, Jelinek W, Darnell JE, et al. Methyl labeling of HeLa cell hnRNA:a comparison with mRNA[J]. Cell, 1976,7(2):227-237.
pmid: 954080 |
| [41] |
Feng Z, Li Q, Meng R, et al. METTL3 regulates alternative splicing of MyD88 upon the lipopolysaccharide-induced inflammatory response in human dental pulp cells[J]. Journal of Cellular and Molecular Medicine, 2018,22(54):2558-2568.
doi: 10.1111/jcmm.2018.22.issue-5 URL |
| [42] |
Fleith RC, Mears HV, Leong XY, et al. IFIT3 and IFIT2/3 promote IFIT1-mediated translation inhibition by enhancing binding to non-self RNA[J]. Nucleic Acids Research, 2018,46(10):5269-5285.
doi: 10.1093/nar/gky191 URL |
| [43] |
Huang H, Weng H, Sun W, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation[J]. Nature Cell Biology, 2018,20(3):285-295.
doi: 10.1038/s41556-018-0045-z URL |
| [44] |
Zhang Z, Chen LQ, Zhao YL, et al. Single-base mapping of m6A by an antibody-independent method[J]. Science Advances, 2019, 5(7):eaax0250.
doi: 10.1126/sciadv.aax0250 URL |
| [45] |
Garcia-Campos MA, Edelheit S, Toth U, et al. Deciphering the “m6A Code” via antibody-independent quantitative profiling[J]. Cell, 2019, 178(3):731-747. e16.
doi: S0092-8674(19)30676-2 pmid: 31257032 |
| [46] |
Ortega A, Niksic M, Bachi A, et al. Biochemical function of female-lethal(2)D/wilms’ tumor suppressor-1-associated proteins in alternative pre-mRNA splicing[J]. Journal of Biological Chemistry, 2003,278(5):3040-3047.
doi: 10.1074/jbc.M210737200 URL |
| [47] |
Batista PJ, Molinie B, Wang J, et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells[J]. Cell Stem Cell, 2014,15(6):707-719.
doi: 10.1016/j.stem.2014.09.019 pmid: 25456834 |
| [48] |
Xiao S, Cao S, Huang Q, et al. The RNA N6-methyladenosine modification landscape of human fetal tissues[J]. Nature Cell Biology, 2019,21(5):651-661.
doi: 10.1038/s41556-019-0315-4 URL |
| [49] |
Batista PJ, Ruby JG, Claycomb JM, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans[J]. Molecular Cell, 2008,31(1):67-78.
doi: 10.1016/j.molcel.2008.06.002 pmid: 18571452 |
| [50] |
Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation[J]. Science, 2015,347(6225):1002-1006.
doi: 10.1126/science.1261417 URL |
| [51] |
Deangelis AM, Martini AE, Owen CM. Assisted reproductive tech-nology and epigenetics[J]. Semin Reprod Med, 2018,36(3/4):221-232.
doi: 10.1055/s-0038-1675780 URL |
| [52] |
Lin Z, Hsu PJ, Xing X, et al. Mettl3-/Mettl14-mediated mRNA N6-methyladenosine modulates murine spermatogenesis[J]. Cell Research, 2017,27(10):1216-1230.
doi: 10.1038/cr.2017.117 URL |
| [53] |
Xu K, Yang Y, Feng GH, et al. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation[J]. Cell Research, 2017,27(9):1100-1114.
doi: 10.1038/cr.2017.100 URL |
| [54] |
Luo Z, Zhang Z, Tai L, et al. Comprehensive analysis of differences of N6-methyladenosine RNA methylomes between high-fat-fed and normal mouse livers[J]. Epigenomics, 2019,11:1267-1282.
doi: 10.2217/epi-2019-0009 URL |
| [55] |
Zhao YL, Liu YH, Wu RF, et al. Understanding m6A function through uncovering the diversity roles of YTH domain-containing proteins[J]. Molecular Biotechnology, 2019,61(5):355-364.
doi: 10.1007/s12033-018-00149-z pmid: 30637606 |
| [56] |
Wang X, Sun B, Jiang Q, et al. mRNA m6A plays opposite role in regulating UCP2 and PNPLA2 protein expression in adipocytes[J]. Int J Obes(Lond), 2018,42(11):1912-1924.
doi: 10.1038/s41366-018-0027-z URL |
| [57] | Wang X, Wu R, Liu Y, et al. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7[J]. Autophagy, 2019: 1-15. |
| [58] |
Wu Y, Xie L, Wang M, et al. Mettl3-mediated m6A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis[J]. Nat Commun, 2018,9(1):4772.
doi: 10.1038/s41467-018-06898-4 URL |
| [59] | Li X, Yang J, Zhu Y, et al. Mouse maternal high-fat intake dynamically programmed mRNA m6A modifications in adipose and skeletal muscle tissues in offspring[J]. Int J Mol Sci, 2016,17(8). |
| [60] |
Liu J, Luo G, Sun J, et al. METTL14 is essential for β-cell survival and insulin secretion[J]. Biochim Biophys Acta Mol Basis Dis, 2019,1865(9):2138-2148.
doi: 10.1016/j.bbadis.2019.04.011 URL |
| [61] | Kudou K, Komatsu T, Nogami J, et al. The requirement of Mettl3-promoted MyoD mRNA maintenance in proliferative myoblasts for skeletal muscle differentiation[J]. Open Biology, 7(9):170119. |
| [62] | Chen JN, Chen Y, Wei YY, et al. Regulation of m6A RNA methylation and its effect on myogenic differentiation in murine myoblasts[J]. Mol Biol(Mosk), 2019,53(3):436-445. |
| [63] |
Wang X, Huang N, Yang M, et al. FTO is required for myogenesis by positively regulating mTOR-PGC-1alpha pathway-mediated mitochondria biogenesis[J]. Cell Death Dis, 2017,8(3):e2702.
doi: 10.1038/cddis.2017.122 URL |
| [64] |
Mathiyalagan P, Adamiak M, Mayourian J, et al. FTO-dependent N6-methyladenosine regulates cardiac function during remodeling and repair[J]. Circulation, 2019,139(4):518-532.
doi: 10.1161/CIRCULATIONAHA.118.033794 pmid: 29997116 |
| [65] |
Dorn LE, Lasman L, Chen J, et al. The N6-Methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy[J]. Circulation, 2019,139(4):533-545.
doi: 10.1161/CIRCULATIONAHA.118.036146 URL |
| [66] |
Kmietczyk V, Riechert E, Kalinski L, et al. m6A-mRNA methylation regulates cardiac gene expression and cellular growth[J]. Life Science Alliance, 2019,2(2):e201800233.
doi: 10.26508/lsa.201800233 URL |
| [67] | Lin J, Zhu Q, Huang J, et al. Hypoxia promotes vascular smooth muscle cell(VSMC)differentiation of adipose-derived stem cell(ADSC)by regulating mettl3 and paracrine factors[J]. Stem Cells International, 2020,2020:2830565. |
| [1] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [2] | 薛宁, 王瑾, 李世新, 刘叶, 程海娇, 张玥, 毛雨丰, 王猛. 多基因同步调控结合高通量筛选构建高产L-苯丙氨酸的谷氨酸棒杆菌工程菌株[J]. 生物技术通报, 2023, 39(9): 268-280. |
| [3] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [4] | 张道磊, 甘雨军, 乐亮, 普莉. 玉米产量性状的表观遗传调控机制和育种应用[J]. 生物技术通报, 2023, 39(8): 31-42. |
| [5] | 叶云芳, 田清尹, 施婷婷, 王亮, 岳远征, 杨秀莲, 王良桂. 植物中β-紫罗兰酮生物合成及调控研究进展[J]. 生物技术通报, 2023, 39(8): 91-105. |
| [6] | 李英, 岳祥华. DNA甲基化在解析毛竹自然变异中的应用[J]. 生物技术通报, 2023, 39(7): 48-55. |
| [7] | 成婷, 苑帅, 张晓元, 林良才, 李欣, 张翠英. 酿酒酵母异丁醇合成途径调控的研究进展[J]. 生物技术通报, 2023, 39(7): 80-90. |
| [8] | 王兵, 赵会纳, 余婧, 余世洲, 雷波. 植物侧枝发育的调控研究进展[J]. 生物技术通报, 2023, 39(5): 14-22. |
| [9] | 史建磊, 宰文珊, 苏世闻, 付存念, 熊自立. 番茄青枯病抗性相关miRNA的鉴定与表达分析[J]. 生物技术通报, 2023, 39(5): 233-242. |
| [10] | 周定定, 李辉虎, 汤兴涌, 余发新, 孔丹宇, 刘毅. 甘草酸和甘草苷生物合成与调控的研究进展[J]. 生物技术通报, 2023, 39(5): 44-53. |
| [11] | 刘晓燕, 祝振亮, 史广宇, 华梓宇, 杨晨, 张涌, 刘军. 乳腺生物反应器的表达优化策略[J]. 生物技术通报, 2023, 39(5): 77-91. |
| [12] | 薛皦, 朱庆锋, 冯彦钊, 陈沛, 刘文华, 张爱霞, 刘勤坚, 张琪, 于洋. 植物基因上游开放阅读框的研究进展[J]. 生物技术通报, 2023, 39(4): 157-165. |
| [13] | 魏明, 王欣玉, 伍国强, 赵萌. NAD依赖型去乙酰化酶SRT在植物表观遗传调控中的作用[J]. 生物技术通报, 2023, 39(4): 59-70. |
| [14] | 陈强, 邹明康, 宋家敏, 张冲, 吴隆坤. 甜瓜LBD基因家族的鉴定和果实发育进程中的表达分析[J]. 生物技术通报, 2023, 39(3): 176-183. |
| [15] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||