








生物技术通报 ›› 2022, Vol. 38 ›› Issue (7): 62-69.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1592
石佳( ), 朱秀梅, 薛梦雨, 余超, 魏一鸣, 杨凤环, 陈华民(
), 朱秀梅, 薛梦雨, 余超, 魏一鸣, 杨凤环, 陈华民( )
)
收稿日期:2021-12-25
出版日期:2022-07-26
发布日期:2022-08-09
作者简介:石佳,女,硕士研究生,研究方向:分子植物病理学;E-mail: 基金资助:
SHI Jia( ), ZHU Xiu-mei, XUE Meng-yu, YU Chao, WEI Yi-ming, YANG Feng-huan, CHEN Hua-min(
), ZHU Xiu-mei, XUE Meng-yu, YU Chao, WEI Yi-ming, YANG Feng-huan, CHEN Hua-min( )
)
Received:2021-12-25
Published:2022-07-26
Online:2022-08-09
摘要:
植物转录因子是植物体内调节基因表达的重要蛋白,参与多种生物学功能的调控。研究植物转录因子调控靶基因的常用方法为染色质免疫共沉淀法(chromatin immunoprecipitation,ChIP),但抗体特异性、遗传材料构建的耗时性等因素却大大限制了ChIP技术的应用范围和效果。本文通过对水稻原生质体瞬时转化、甲醛固定、免疫沉淀核酸的超声波破碎等条件的优化,建立了基于水稻原生质体的染色质免疫共沉淀技术体系(chromatin immunoprecipitation system based on rice protoplasts,ChIP-RP);并通过本技术体系验证了水稻转录因子OsNF-YA4蛋白对靶基因序列的富集作用。本技术体系将减少制备特异性抗体或构建稳定遗传材料的局限,有利于水稻转录因子直接调控靶基因的快速筛选和验证,推动水稻转录因子调控功能的分子机制解析。
石佳, 朱秀梅, 薛梦雨, 余超, 魏一鸣, 杨凤环, 陈华民. 基于水稻原生质体的染色质免疫共沉淀技术优化及应用[J]. 生物技术通报, 2022, 38(7): 62-69.
SHI Jia, ZHU Xiu-mei, XUE Meng-yu, YU Chao, WEI Yi-ming, YANG Feng-huan, CHEN Hua-min. Optimization and Application of the Chromatin Immunoprecipitation Based on Rice Protoplast[J]. Biotechnology Bulletin, 2022, 38(7): 62-69.
| 家族Family | 基因Gene | ID | CCAAT 数目及位置CCAAT number and position |
|---|---|---|---|
| OsNRT1家族(低亲和力)OsNRT1 family(low affinity) | OsNRT1.1 | Os03g13274 | 2(-1404,-1055) |
| OsNRT2家族(高亲和力)OsNRT2 family(high affinity) | OsNRT2.1 | Os02g02170 | 1(-175) |
| OsNRT2.2 | Os02g02190 | 2(-276,-53) | |
| OsNAR2.1 | Os02g38230 | 2(-631,-460) |
表1 NRTs基因启动子区域调控元件的预测
Table 1 Prediction of regulatory elements in the promoter region of NRTs genes
| 家族Family | 基因Gene | ID | CCAAT 数目及位置CCAAT number and position |
|---|---|---|---|
| OsNRT1家族(低亲和力)OsNRT1 family(low affinity) | OsNRT1.1 | Os03g13274 | 2(-1404,-1055) |
| OsNRT2家族(高亲和力)OsNRT2 family(high affinity) | OsNRT2.1 | Os02g02170 | 1(-175) |
| OsNRT2.2 | Os02g02190 | 2(-276,-53) | |
| OsNAR2.1 | Os02g38230 | 2(-631,-460) |
| 基因名称 Gene name | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 片段大小及内容 Segment size and content |
|---|---|---|---|
| OsNRT1.1 | OsNRT1.1 Q-F2 | TGCTACGGTCTCATCTTCTCT | 199 bp 包含NRT1.1基因启动子序列中的第二个CCAAT-box 199 bp contains the second CCAAT-box in the promoter of NRT1.1 |
| OsNRT1.1 Q-R2 | CAAGTAATCCATCTAACGCTACGA | ||
| OsNRT2.1 | OsNRT2.1 Q-F1 | ACGAATCTTAAGGCAAAT | 194 bp包含NRT2.1基因启动子序列中的一个CCAAT-box 199 bp contains a CCAAT-box in the promoter of NRT2.1 |
| OsNRT2.1 Q-R1 | CTTCTTGGAGATGGAATC | ||
| OsNRT2.2 | OsNRT2.2 Q-F2 | CACGAGGCAGATATTACAACTTGA | 149 bp 包含NRT2.2基因启动子序列中的第二个CCAAT-box 149 bp contains the second CCAAT-box in the promoter of NRT2.2 |
| OsNRT2.2 Q-R2 | CGGTGACGATGATCTTGGC | ||
| OsNAR2.1 | OsNAR2.1 Q-F2 | TTCCTCCATTAAGAACGCCTTC | 119 bp 包含NAR2.1基因启动子序列中的第二个CCAAT-box 119 bp contains the second CCAAT-box in the promoter of NAR2.1 |
| OsNAR2.1 Q-R2 | TTGAGTGCCTCGGTTGTTG | ||
| Negative | OsNAR2.1-NF | GATGGCTGTCCTGCTCTTG | 183 bp 为NAR2.1基因启动子中不含CCAAT-box的序列 183 bp NAR2.1 gene promoter without CCAAT-box |
| OsNAR2.1-NR | ATCATTTCGCTCCTCCAAACTATT |
表2 RT-qPCR验证所需的引物序列
Table 2 Required primer sequences for RT-qPCR validation
| 基因名称 Gene name | 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 片段大小及内容 Segment size and content |
|---|---|---|---|
| OsNRT1.1 | OsNRT1.1 Q-F2 | TGCTACGGTCTCATCTTCTCT | 199 bp 包含NRT1.1基因启动子序列中的第二个CCAAT-box 199 bp contains the second CCAAT-box in the promoter of NRT1.1 |
| OsNRT1.1 Q-R2 | CAAGTAATCCATCTAACGCTACGA | ||
| OsNRT2.1 | OsNRT2.1 Q-F1 | ACGAATCTTAAGGCAAAT | 194 bp包含NRT2.1基因启动子序列中的一个CCAAT-box 199 bp contains a CCAAT-box in the promoter of NRT2.1 |
| OsNRT2.1 Q-R1 | CTTCTTGGAGATGGAATC | ||
| OsNRT2.2 | OsNRT2.2 Q-F2 | CACGAGGCAGATATTACAACTTGA | 149 bp 包含NRT2.2基因启动子序列中的第二个CCAAT-box 149 bp contains the second CCAAT-box in the promoter of NRT2.2 |
| OsNRT2.2 Q-R2 | CGGTGACGATGATCTTGGC | ||
| OsNAR2.1 | OsNAR2.1 Q-F2 | TTCCTCCATTAAGAACGCCTTC | 119 bp 包含NAR2.1基因启动子序列中的第二个CCAAT-box 119 bp contains the second CCAAT-box in the promoter of NAR2.1 |
| OsNAR2.1 Q-R2 | TTGAGTGCCTCGGTTGTTG | ||
| Negative | OsNAR2.1-NF | GATGGCTGTCCTGCTCTTG | 183 bp 为NAR2.1基因启动子中不含CCAAT-box的序列 183 bp NAR2.1 gene promoter without CCAAT-box |
| OsNAR2.1-NR | ATCATTTCGCTCCTCCAAACTATT |
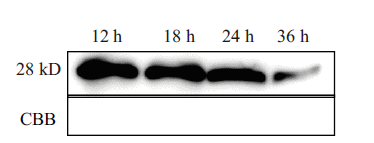
图2 原生质体转化后OsNF-YA4蛋白表达水平的检测 表达载体转化水稻原生质体后,孵育不同时间,提取蛋白,用anti-Myc抗体检测融合蛋白的表达量 1-4:分别为12 h,18 h,24 h,36 h时NF-YA4蛋白表达水平;考马斯亮蓝染色显示各个泳道蛋白上样量
Fig.2 Expressions detection of OsNF-YA4 protein after protoplast transformed After rice protoplast transformed with expression vector and the incubation was performed at different times,the transient expressed proteins were extracted,and the expression of fused protein was detected via anti-Myc antibody 1-4:NF-YA4 protein expressions at 12,18,24 and 36 h. Coomassie brilliant blue staining showed the loaded protein in each lane
| 甲醛终浓度 Final concentration of formaldehyde/% | 细胞活性百分比 Percentage of cell activity/% |
|---|---|
| 0 | 100+0.01 |
| 0.1 | 93+2.79 |
| 0.3 | 80+4.27 |
| 0.7 | 72+3.98 |
| 1.0 | 48+2.72 |
表3 不同甲醛终浓度对原生质体细胞活性的影响
Table 3 Effects of different final concentrations of formal-dehyde on protoplasmic cell activity
| 甲醛终浓度 Final concentration of formaldehyde/% | 细胞活性百分比 Percentage of cell activity/% |
|---|---|
| 0 | 100+0.01 |
| 0.1 | 93+2.79 |
| 0.3 | 80+4.27 |
| 0.7 | 72+3.98 |
| 1.0 | 48+2.72 |
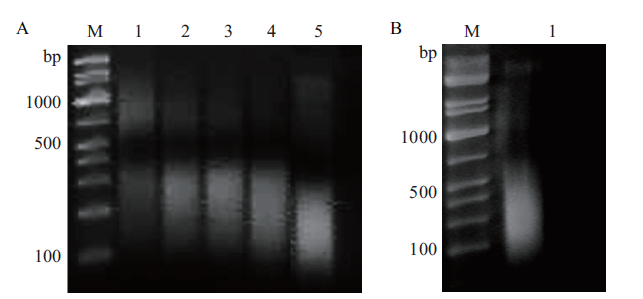
图3 OsNF-YA4蛋白ChIPs富集DNA的超声破碎时间优化(A)和效果检测(B) A:超声破碎时长的效果比较(M:DL2000 marker;1-5:超声时长分别为5 min,10 min,15 min,20 min,30 min);B:ChIPs富集DNA的破碎效果(1:超声时长为15 min时,input样品的破碎效果)
Fig.3 Optimization of ultrasonic fragmentation time on OsNF-YA4 ChIPs-enriched DNA(A)and validation of the fragmentation(B) A:Effects of ultrasonic crushing time on fragment length(M:DL2000 marker;1-5:ultrasonic duration was 5,10,15,20 and 30 min respectivey)B:The fragmentation of DNA enrichment by ChIPs(1:the fragmentation of input when ultrasonic treated 15 min)
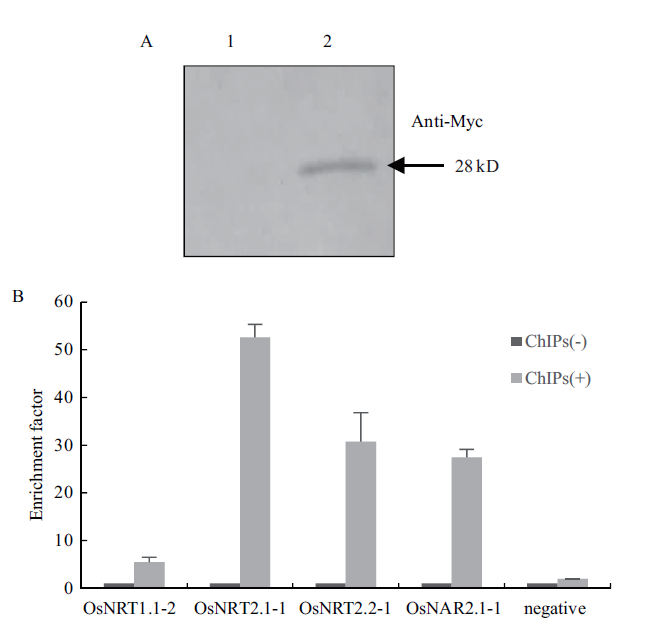
图4 染色质免疫共沉淀效果的检测(A)和所富集DNA的特异性分析(B) A:Western blot检测免疫沉淀后目的蛋白OsNFYA4的水平(1:不加anti-Myc抗体的免疫沉淀样品(ChIPs-);2:加anti-Myc抗体的免疫沉淀样品(ChIPs+));B:qRT-PCR检测免疫沉淀后所富集DNA的特异性
Fig.4 Detection of the effect of chromosome immunopreci-pitation(A)and specificity analysis of enriched DNA(B)A:The level of OsNFYA4 after immunoprecipitation detected by Western blot(1:immunoprecipitated samples(ChIPs-)without anti-Myc antibody;2:immunoprecipitated samples with anti-Myc antibody(ChIPs+)). B:Validation of the specificity of the ChIP-RP enriched DNA by qRT-PCR
| [1] |
Mitsuda N, Ohme-Takagi M. Functional analysis of transcription factors in Arabidopsis[J]. Plant Cell Physiol, 2009, 50(7):1232-1248.
doi: 10.1093/pcp/pcp075 URL |
| [2] |
Zou YM, Wang SF, Zhou YY, et al. Transcriptional regulation of the immune receptor FLS2 controls the ontogeny of plant innate immunity[J]. Plant Cell, 2018, 30(11):2779-2794.
doi: 10.1105/tpc.18.00297 URL |
| [3] |
Bannister AJ, Kouzarides T. Basic peptides enhance protein/DNA interaction in vitro[J]. Nucleic Acids Res, 1992, 20(13):3523.
pmid: 1630932 |
| [4] |
Wang MM, Reed RR. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast[J]. Nature, 1993, 364(6433):121-126.
doi: 10.1038/364121a0 URL |
| [5] |
杨立文, 刘双荣, 李玉红, 等. 植物转录因子与DNA互作研究技术[J]. 植物学报, 2020, 55(4):468-474.
doi: 10.11983/CBB20057 |
| Yang LW, Liu SR, Li YH, et al. Methods for examining transcription factor-DNA interaction in plants[J]. Chin Bull Bot, 2020, 55(4):468-474. | |
| [6] |
Gilmour DS, Lis JT. Detecting protein-DNA interactions in vivo:distribution of RNA polymerase on specific bacterial genes[J]. Proc Natl Acad Sci USA, 1984, 81(14):4275-4279.
doi: 10.1073/pnas.81.14.4275 URL |
| [7] |
Solomon MJ, Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking:a probe for in vivo chromatin structures[J]. Proc Natl Acad Sci USA, 1985, 82(19):6470-6474.
doi: 10.1073/pnas.82.19.6470 URL |
| [8] |
Mundade R, Ozer HG, Wei H, et al. Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond[J]. Cell Cycle, 2014, 13(18):2847-2852.
doi: 10.4161/15384101.2014.949201 pmid: 25486472 |
| [9] |
Kaufmann K, Muiño JM, Østerås M, et al. Chromatin immunoprecipitation(ChIP)of plant transcription factors followed by sequencing(ChIP-SEQ)or hybridization to whole genome arrays(ChIP-CHIP)[J]. Nat Protoc, 2010, 5(3):457-472.
doi: 10.1038/nprot.2009.244 pmid: 20203663 |
| [10] |
Nardini M, Gnesutta N, Donati G, et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination[J]. Cell, 2013, 152(1/2):132-143.
doi: 10.1016/j.cell.2012.11.047 URL |
| [11] |
Zhao BT, Ge LF, Liang RQ, et al. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor[J]. BMC Mol Biol, 2009, 10:29.
doi: 10.1186/1471-2199-10-29 URL |
| [12] |
Luan MD, Xu MY, Lu YM, et al. Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves[J]. Gene, 2015, 555(2):178-185.
doi: 10.1016/j.gene.2014.11.001 URL |
| [13] |
Ding Q, Zeng J, He XQ. MiR169 and its target PagHAP2-6 regulated by ABA are involved in poplar cambium dormancy[J]. J Plant Physiol, 2016, 198:1-9.
doi: 10.1016/j.jplph.2016.03.017 URL |
| [14] |
Li WX, Oono Y, Zhu JH, et al. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance[J]. Plant Cell, 2008, 20(8):2238-2251.
doi: 10.1105/tpc.108.059444 URL |
| [15] |
Zhao M, Ding H, Zhu JK, et al. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis[J]. New Phytol, 2011, 190(4):906-915.
doi: 10.1111/j.1469-8137.2011.03647.x URL |
| [16] |
Combier JP, Frugier F, de Billy F, et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula[J]. Genes Dev, 2006, 20(22):3084-3088.
doi: 10.1101/gad.402806 URL |
| [17] |
Warpeha KM, Upadhyay S, Yeh J, et al. The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis[J]. Plant Physiol, 2007, 143(4):1590-1600.
doi: 10.1104/pp.106.089904 URL |
| [18] |
Xu MY, Zhang L, Li WW, et al. Stress-induced early flowering is mediated by miR169 in Arabidopsis thaliana[J]. J Exp Bot, 2013, 65(1):89-101.
doi: 10.1093/jxb/ert353 URL |
| [19] | Para A, Li Y, Coruzzi GM. μChIP-seq for genome-wide mapping of in vivo TF-DNA interactions in Arabidopsis root protoplasts[J]. Methods Mol Biol Clifton N J, 2018, 1761:249-261. |
| [20] | He F, Zhang F, Sun WX, et al. A versatile vector toolkit for functional analysis of rice genes[J]. Rice(N Y), 2018, 11(1):27. |
| [21] | Chen HM, Zou Y, Shang YL, et al. Firefly luciferase complementation imaging assay for protein-protein interactions in plants[J]. Plant Physiol, 2008, 146(2):368-376. |
| [22] |
Yu C, Chen YT, Cao YQ, et al. Overexpression of miR169o, an overlapping microRNA in response to both nitrogen limitation and bacterial infection, promotes nitrogen use efficiency and susceptibility to bacterial blight in rice[J]. Plant Cell Physiol, 2018, 59(6):1234-1247.
doi: 10.1093/pcp/pcy060 URL |
| [23] | 翁小煜, 周少立, 宗伟, 等. 水稻染色质免疫共沉淀[M]. Bio-protocol, 2018, Bio-101:e1010135. |
|
Weng XY, Zhou SL, Zong W, et al. ChIP assay in rice[M]. Bio-protocol, 2018, Bio-101:e1010135. DOI: 10.21769/BioProtoc.1010135.
doi: 10.21769/BioProtoc.1010135 |
|
| [24] |
Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant Cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Res, 2002, 30(1):325-327.
doi: 10.1093/nar/30.1.325 URL |
| [25] |
Baranello L, Kouzine F, Sanford S, et al. ChIP bias as a function of cross-linking time[J]. Chromosome Res, 2016, 24(2):175-181.
doi: 10.1007/s10577-015-9509-1 pmid: 26685864 |
| [26] |
Ricardi MM, González RM, Iusem ND. Protocol:fine-tuning of a Chromatin Immunoprecipitation(ChIP)protocol in tomato[J]. Plant Methods, 2010, 6:11.
doi: 10.1186/1746-4811-6-11 URL |
| [27] | 董浩, 彭小薇, 王晓英, 等. 染色质免疫共沉淀技术对羊种布鲁氏菌转录调控因子MucR靶基因的筛选[J]. 中国农业大学学报, 2016, 21(4):102-106. |
| Dong H, Peng XW, Wang XY, et al. Identification of the targets of MucR by chromatin immunoprecipitation in Brucella melitensis[J]. J China Agric Univ, 2016, 21(4):102-106. |
| [1] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [2] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [3] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [4] | 徐靖, 朱红林, 林延慧, 唐力琼, 唐清杰, 王效宁. 甘薯IbHQT1启动子的克隆及上游调控因子的鉴定[J]. 生物技术通报, 2023, 39(8): 213-219. |
| [5] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [6] | 陈晓, 于茗兰, 吴隆坤, 郑晓明, 逄洪波. 植物lncRNA及其对低温胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(7): 1-12. |
| [7] | 郭怡婷, 赵文菊, 任延靖, 赵孟良. 菊芋NAC转录因子家族基因的鉴定及分析[J]. 生物技术通报, 2023, 39(6): 217-232. |
| [8] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [9] | 王兵, 赵会纳, 余婧, 余世洲, 雷波. 植物侧枝发育的调控研究进展[J]. 生物技术通报, 2023, 39(5): 14-22. |
| [10] | 张新博, 崔浩亮, 史佩华, 高锦春, 赵顺然, 陶晨雨. 低起始量的免疫共沉淀技术研究进展[J]. 生物技术通报, 2023, 39(4): 227-235. |
| [11] | 葛颜锐, 赵冉, 徐静, 李若凡, 胡云涛, 李瑞丽. 植物维管形成层发育及其调控的研究进展[J]. 生物技术通报, 2023, 39(3): 13-25. |
| [12] | 刘铖霞, 孙宗艳, 罗云波, 朱鸿亮, 曲桂芹. bHLH转录因子的磷酸化调控植物生理功能的研究进展[J]. 生物技术通报, 2023, 39(3): 26-34. |
| [13] | 赵孟良, 郭怡婷, 任延靖. 菊芋WRKY转录因子家族基因的鉴定及分析[J]. 生物技术通报, 2023, 39(2): 116-125. |
| [14] | 韩芳英, 胡昕, 王楠楠, 谢裕红, 王晓艳, 朱强. DREBs响应植物非生物逆境胁迫研究进展[J]. 生物技术通报, 2023, 39(11): 86-98. |
| [15] | 陈楚怡, 杨小梅, 陈胜艳, 陈斌, 岳莉然. ABA和干旱胁迫下菊花脑ZF-HD基因家族的表达分析[J]. 生物技术通报, 2023, 39(11): 270-282. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||