








生物技术通报 ›› 2023, Vol. 39 ›› Issue (3): 81-88.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0577
蔡梦鲜( ), 高作敏, 胡利娟, 冯群, 王洪程, 朱斌(
), 高作敏, 胡利娟, 冯群, 王洪程, 朱斌( )
)
收稿日期:2022-05-10
出版日期:2023-03-26
发布日期:2023-04-10
通讯作者:
朱斌,男,博士,教授,硕士生导师,研究方向:油菜遗传育种;E-mail: zhugg130@126.com作者简介:蔡梦鲜,女,硕士,研究方向:油菜遗传育种;E-mail: caimengxian@126.com
基金资助:
CAI Meng-xian( ), GAO Zuo-min, HU Li-juan, FENG Qun, WANG Hong-cheng, ZHU Bin(
), GAO Zuo-min, HU Li-juan, FENG Qun, WANG Hong-cheng, ZHU Bin( )
)
Received:2022-05-10
Published:2023-03-26
Online:2023-04-10
摘要:
缺体(nullisomic,2(n-1))是相对于正常生物个体(euploid,2n)的染色体组缺失一对同源染色体的有机体,是一种不可多得的遗传材料,能够实现对单条染色体的遗传解析。本研究通过甘蓝型油菜(Brassica napus,2n=38,AnAnCnCn)与先前获得的剥离白菜(restituted B. rapa,RBR,2n=20,AnAn)进行杂交及连续回交,通过C基因组染色体特异分子标记及荧光原位杂交(flourescence in situ hybridization,FISH)技术首次筛选出了甘蓝型油菜C染色体组的两个缺体系:C1染色体(NC1)和C2染色体缺失的缺体系(NC2)。随后对两个缺体材料的花粉母细胞(pollen mathor cell,PMC)观察证实,在终变期的PMC呈现18个二价体,减I后期染色体以18:18均等分离为主,但在后期I及后期Ⅱ均出现不同频率的落后染色体。与亲本相比,两个缺体的生物量、花粉育性、结实率等显著降低,且表现一些特有性状,例如NC1全叶被覆毛刺,NC2开花时间比亲本提早近两个月,说明缺失染色体上携带有相关性状的抑制基因。这两个缺体材料有效降低了甘蓝型油菜基因组的复杂性,有助于目标性状的初步定位,或者基因定位结果的验证,为今后的甘蓝型油菜遗传研究提供便利。
蔡梦鲜, 高作敏, 胡利娟, 冯群, 王洪程, 朱斌. 天然甘蓝型油菜C染色体组C1,C2缺体的创建及遗传分析[J]. 生物技术通报, 2023, 39(3): 81-88.
CAI Meng-xian, GAO Zuo-min, HU Li-juan, FENG Qun, WANG Hong-cheng, ZHU Bin. Development and Genetic Analysis of Two Nullisomic Lines(NC1 and NC2)in Natural Brassica napus[J]. Biotechnology Bulletin, 2023, 39(3): 81-88.
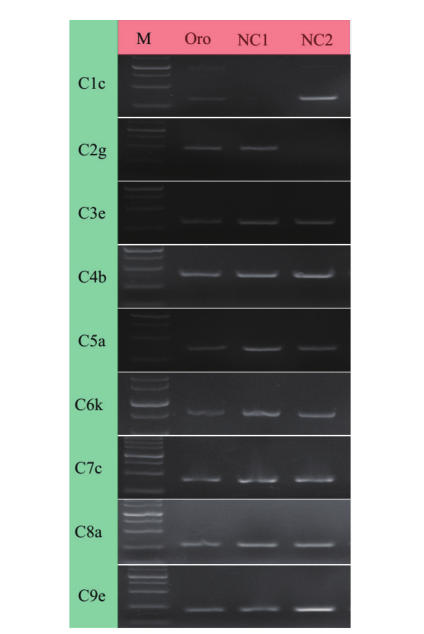
图2 NC1和NC2缺体单株的C染色体特异分子标记鉴定结果 M:Marker标记大小分别为2 000、1 500、1 000、750、500、250、100 bp;Oro是甘蓝型油菜;NC1,NC2代表甘蓝型油菜C染色体缺体;C1c,C2g,…C9e为C基因组染色体特异性引物
Fig. 2 Identification of C chromosome nullisomic-specific markers in NC1 and NC2 M: Marker. The mark sizes were 2 000, 1 500, 1 000, 750, 500, 250 and 100 bp. Oro is B. napus. NC1 and NC2 refer to the nullisomic of chromosome C in B. napus. C1c, C2g, …C9e are chromosomal specific primers for C genome
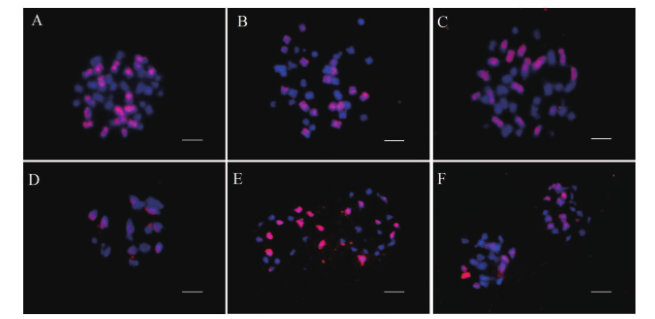
图3 甘蓝型油菜“ Oro ”,NC1及NC2的体细胞及花粉母细胞荧光原位杂交分析 A:“Oro”有丝分裂细胞,有38条染色体(蓝色),包括18条C亚基因组染色体(红色);B-C:NC1,NC2有丝分裂细胞,有36条染色体(蓝色),包括16条C亚基因组染色体(红色);D:C2缺体终变期,显示8个红色杂交信号;E-F:NC1及NC2中处于减数分裂后I期的PMC检测到16个杂交信号均等分离。蓝色表示DAPI染色,红色表示BAC BoB014O06探针信号。标尺=10 μm
Fig. 3 FISH analysis of somatic cells and pollen mother cells of B. napus “Oro”,NC1,and NC2 A: “Oro” mitotic cells with 38 chromosomes(blue), including 18 C subgenomic chromosomes(red). B-C: NC1, NC2 mitotic cells with 36 chromosomes(blue), including 16 C subgenomic chromosomes(red). D: C2 showing 8 red hybridization signals at diakinesis. E-F: The PMC of NC2 and NC1 show equal separation of 16 hybridization signals at anaphase I. Blue indicates DAPI staining and red indicates BAC BoB014O06 probe signal. Bar: 10 μm

图4 亲本甘蓝型油菜“Oro”,NC1,NC2表型及花粉育性 A:亲本甘蓝型油菜“Oro”;B:NC1植株,放大框中显示NC1叶脉、叶片及叶边缘被覆白色毛刺;C:NC2植株,放大框中显示NC2已现蕾;E-F:“Oro”,NC1和NC2的花粉育性
Fig. 4 Phenotype and pollen fertility of parental B. napus “Oro”,NC1,and NC2 A: Parental B. napus “Oro”. B: NC1 plants. The enlargement shows that blade, vein, and leaf margin cover with burrs. C: NC2 plants. The enlargement shows that NC2 plant has early squaring stage. E-F: The pollen fertility of “Oro”, NC1, and NC2

图5 甘蓝型油菜NC1与 NC2减数分裂染色体行为观察 A-D:依次为NC1体细胞染色体数(2n=36),花粉母细胞减数DK、AP I及AP II时期的细胞;E-H:依次为NC2体细胞染色体数(2n=36),花粉母细胞减数DK、AP I及AP II时期的细胞;I:正常配对的缺体中I期细胞;J,K:NC1及NC2中出现染色体滞后现象的后I期细胞;L:NC2中AP II时期出现滞后染色体的PMC。箭头所指为落后染色体,标尺=10 μm
Fig. 5 Observation of chromosome behavior of at meiosis of NC1 and NC2 in B. napus A-D: Somatic chromosome number(2n=36)of NC1, PMCs of NC1 at DK, AP I, and AP II, respectively. E-H: Somatic chromosome number(2n=36)of NC2, PMCs of NC2 at DK, AP I, and AP II, respectively. I: the PMC of nullisomic show normal pairing at metaphase I. J, K: The PMCs of NC1 and NC2 with delaying chromosome at AP I. L: The PMC of NC2 with delaying chromosome at AP II. The arrows show the delaying chromosomes. Bar: 10 μm
| [1] |
Siegel JJ, Amon A. New insights into the troubles of aneuploidy[J]. Annu Rev Cell Dev Biol, 2012, 28:189-214.
doi: 10.1146/annurev-cellbio-101011-155807 pmid: 22804579 |
| [2] | Sears ER. Nullisomic-tetrasomic combinations in hexaploid wheat[M]//Riley R, Lewis KR. Chromosome manipulations and plant genetics. Boston: Springer, 1966:29-45. |
| [3] | Zhu B, Shao YJ, Pan Q, et al. Genome-wide gene expression perturbation induced by loss of C2 chromosome in allotetraploid Brassica napus L[J]. Front Plant Sci, 2015, 6:763. |
| [4] |
Wang M, Wang SB, Liang Z, et al. From genetic stock to genome editing:gene exploitation in wheat[J]. Trends Biotechnol, 2018, 36(2):160-172.
doi: 10.1016/j.tibtech.2017.10.002 URL |
| [5] |
Francki MG, Hayton S, Gummer JPA, et al. Metabolomic profiling and genomic analysis of wheat aneuploid lines to identify genes controlling biochemical pathways in mature grain[J]. Plant Biotechnol J, 2016, 14(2):649-660.
doi: 10.1111/pbi.12410 pmid: 26032167 |
| [6] |
Liu M, Rathjen T, Weligama K, et al. Analysis of aneuploid lines of bread wheat to map chromosomal locations of genes controlling root hair length[J]. Ann Bot, 2017, 119(8):1333-1341.
doi: 10.1093/aob/mcx030 URL |
| [7] |
Li TT, Sun YL, Liu TX, et al. TaCER1-1A is involved in cuticular wax alkane biosynthesis in hexaploid wheat and responds to plant abiotic stresses[J]. Plant Cell Environ, 2019, 42(11):3077-3091.
doi: 10.1111/pce.v42.11 URL |
| [8] |
Draeger T, C Martin A, Alabdullah AK, et al. Dmc1 is a candidate for temperature tolerance during wheat meiosis[J]. Theor Appl Genet, 2020, 133(3):809-828.
doi: 10.1007/s00122-019-03508-9 pmid: 31853574 |
| [9] | Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time[J]. Nat Rev Mol Cell Biol, 2007, 8(5):379-393. |
| [10] |
Tu YQ, Sun J, Ge XH, et al. Production and genetic analysis of partial hybrids from intertribal sexual crosses between Brassica napus and Isatis indigotica and progenies[J]. Genome, 2010, 53(2):146-156.
doi: 10.1139/g09-093 pmid: 20140033 |
| [11] |
Zhu B, Tu YQ, Zeng P, et al. Extraction of the constituent subgenomes of the natural allopolyploid rapeseed(Brassica napus L.)[J]. Genetics, 2016, 204(3):1015-1027.
doi: 10.1534/genetics.116.190967 URL |
| [12] |
Chalhoub B, Denoeud F, Liu SY, et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome[J]. Science, 2014, 345(6199):950-953.
doi: 10.1126/science.1253435 pmid: 25146293 |
| [13] |
Xiong ZY, Gaeta RT, Pires JC. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus[J]. Proc Natl Acad Sci USA, 2011, 108(19):7908-7913.
doi: 10.1073/pnas.1014138108 URL |
| [14] |
Li Z, Liu HL, Luo P. Production and cytogenetics of intergeneric hybrids between Brassica napus and Orychophragmus violaceus[J]. Theor Appl Genet, 1995, 91(1):131-136.
doi: 10.1007/BF00220869 pmid: 24169678 |
| [15] |
Zhong XB, Jong JH, Zabel P. Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridization(FISH)[J]. Chromosome Res, 1996, 4(1):24-28.
doi: 10.1007/BF02254940 pmid: 8653264 |
| [16] |
Cui C, Ge XH, Gautam M, et al. Cytoplasmic and genomic effects on meiotic pairing in Brassica hybrids and allotetraploids from pair crosses of three cultivated diploids[J]. Genetics, 2012, 191(3):725-738.
doi: 10.1534/genetics.112.140780 URL |
| [17] | Nayidu NK, Bonham-Smith P, Gruber M. Brassica villosa a potential tool to improve the insect or disease resistance of Brassica crop species[J]. Transcriptomics, 2015, 3(2):114. |
| [18] |
Alahakoon UI, Taheri A, Nayidu NK, et al. Hairy Canola(Brasssica napus)re-visited:down-regulating TTG1 in an AtGL3-enhanced hairy leaf background improves growth, leaf trichome coverage, and metabolite gene expression diversity[J]. BMC Plant Biol, 2016, 16:12.
doi: 10.1186/s12870-015-0680-5 pmid: 26739276 |
| [19] |
Nayidu NK, Kagale S, Taheri A, et al. Comparison of five major trichome regulatory genes in Brassica villosa with orthologues within the Brassicaceae[J]. PLoS One, 2014, 9(4):e95877.
doi: 10.1371/journal.pone.0095877 URL |
| [20] |
Zheng KJ, Tian HN, Hu QN, et al. Ectopic expression of R3 MYB transcription factor gene OsTCL1 in Arabidopsis, but not rice, affects trichome and root hair formation[J]. Sci Rep, 2016, 6:19254.
doi: 10.1038/srep19254 |
| [21] |
Li F, Chen BY, Xu K, et al. A genome-wide association study of plant height and primary branch number in rapeseed(Brassica napus)[J]. Plant Sci, 2016, 242:169-177.
doi: 10.1016/j.plantsci.2015.05.012 URL |
| [22] | Sun CM, Wang BQ, Yan L, et al. Genome-wide association study provides insight into the genetic control of plant height in rapeseed(Brassica napus L.)[J]. Front Plant Sci, 2016, 7:1102. |
| [23] |
Zheng M, Peng C, Liu HF, et al. Genome-wide association study reveals candidate genes for control of plant height, branch initiation height and branch number in rapeseed(Brassica napus L.)[J]. Front Plant Sci, 2017, 8:1246.
doi: 10.3389/fpls.2017.01246 pmid: 28769955 |
| [24] |
Li Z, McKibben MTW, Finch GS, et al. Patterns and processes of diploidization in land plants[J]. Annu Rev Plant Biol, 2021, 72:387-410.
doi: 10.1146/annurev-arplant-050718-100344 pmid: 33684297 |
| [25] |
Griffiths S, Sharp R, Foote TN, et al. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat[J]. Nature, 2006, 439(7077):749-752.
doi: 10.1038/nature04434 |
| [26] |
Serra H, Svačina R, Baumann U, et al. Ph2 encodes the mismatch repair protein MSH7-3D that inhibits wheat homoeologous recombination[J]. Nat Commun, 2021, 12(1):803.
doi: 10.1038/s41467-021-21127-1 |
| [27] |
Higgins EE, Howell EC, Armstrong SJ, et al. A major quantitative trait locus on chromosome A9, BnaPh1, controls homoeologous recombination in Brassica napus[J]. New Phytol, 2021, 229(6):3281-3293.
doi: 10.1111/nph.16986 pmid: 33020949 |
| [1] | 李心怡, 姜春秀, 薛丽, 蒋洪涛, 姚伟, 邓祖湖, 张木清, 余凡. 多荧光标记引物增强甘蔗染色体寡聚核苷酸探针杂交信号[J]. 生物技术通报, 2023, 39(5): 103-111. |
| [2] | 肖小军, 陈明, 韩德鹏, 余跑兰, 郑伟, 肖国滨, 周庆红, 周会汶. 甘蓝型油菜每角果粒数全基因组关联分析[J]. 生物技术通报, 2023, 39(3): 143-151. |
| [3] | 余世洲, 曹领改, 王世泽, 刘勇, 边文杰, 任学良. 烟草种质基因分型核心SNP标记的开发[J]. 生物技术通报, 2023, 39(3): 89-100. |
| [4] | 支添添, 周舟, 陈纪鹏, 韩成云. 甘蓝型油菜酪氨酸代谢关键基因FAH的克隆、功能鉴定和表达分析[J]. 生物技术通报, 2023, 39(10): 115-127. |
| [5] | 金姣姣, 刘自刚, 米文博, 徐明霞, 邹娅, 徐春梅, 赵彩霞. 利用RNA-Seq鉴定调控甘蓝型油菜叶片光合特性的低温胁迫应答基因[J]. 生物技术通报, 2022, 38(4): 126-142. |
| [6] | 郑惠清, 郭仲杰, 蔡志欣, 卢园萍, 廖剑华, 陈美元. 双孢蘑菇野生种质资源营养成分分析与评价[J]. 生物技术通报, 2021, 37(11): 109-118. |
| [7] | 王衍莉, 杨义明, 范书田, 赵滢, 许培磊, 路文鹏, 李昌禹. 基于SSR分子标记的73份山葡萄及杂交后代的遗传多样性分析[J]. 生物技术通报, 2021, 37(1): 189-197. |
| [8] | 孙嘉栋, 孙晓凤, 李兰, 沈伟, 程顺峰. 干细胞技术在地方猪种质资源保护中的应用前景[J]. 生物技术通报, 2020, 36(8): 228-234. |
| [9] | 何硕康, 罗泽伟. QUARTET突变四倍体拟南芥的获得与表型分析[J]. 生物技术通报, 2018, 34(7): 119-125. |
| [10] | 石建斌, 周红, 王宁, 许庆华, 乔文青, 严根土. 棉花SSR标记种质资源纯度鉴定及遗传多样性分析[J]. 生物技术通报, 2018, 34(7): 138-146. |
| [11] | 宋伟凤, 李明聪, 高峥. 环境中微生物原位检测方法研究进展[J]. 生物技术通报, 2017, 33(10): 26-32. |
| [12] | 张彬, 王长利. 端粒长度检测方法及其应用[J]. 生物技术通报, 2016, 32(11): 93-98. |
| [13] | 郝晓云, 沈海涛, 李鸿彬. 甘蓝型油菜下胚轴和带柄子叶再生体系研究[J]. 生物技术通报, 2013, 0(4): 69-74. |
| [14] | 高义平,董福双,王海波. 红小豆生物技术研究进展[J]. 生物技术通报, 2013, 0(3): 10-14. |
| [15] | 张庆田, 范书田, 杨义明, 李晓艳, 宋润刚, 路文鹏. 山葡萄分子生物学研究进展[J]. 生物技术通报, 2013, 0(12): 1-5. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||