








生物技术通报 ›› 2024, Vol. 40 ›› Issue (1): 72-85.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0657
何思成( ), 张紫瑗, 韩雨晴, 苗琳, 张翠英, 于爱群(
), 张紫瑗, 韩雨晴, 苗琳, 张翠英, 于爱群( )
)
收稿日期:2023-07-11
出版日期:2024-01-26
发布日期:2024-02-06
通讯作者:
于爱群,男,博士,教授,研究方向:发酵工程、微生物学、合成生物学;E-mail: yuaiqun@tust.edu.cn作者简介:何思成,男,研究方向:生物工程;E-mail: hesicheng2002@tust.edu.cn
基金资助:
HE Si-cheng( ), ZHANG Zi-yuan, HAN Yu-qing, MIAO Lin, ZHANG Cui-ying, YU Ai-qun(
), ZHANG Zi-yuan, HAN Yu-qing, MIAO Lin, ZHANG Cui-ying, YU Ai-qun( )
)
Received:2023-07-11
Published:2024-01-26
Online:2024-02-06
摘要:
多不饱和脂肪酸属直链脂肪酸,是机体生物膜的关键结构组分,它可以调控糖脂代谢及激素代谢,具有促进发育、提高免疫力和预防疾病等多种益生功能,对机体健康具有十分重要的作用。因此,多不饱和脂肪酸在食品、医药和饲料等多个领域均表现出重要的应用价值和广阔的开发前景,市场需求持续上升。与传统的海洋生物提取法相比,微生物合成法具有生产周期短、工艺简单、对环境友好等优势,近年来利用微生物细胞工厂生产多不饱和脂肪酸等微生物油脂逐渐成为科学界和工业界的研究热点和发展趋势。解脂耶氏酵母作为一种非常规产油酵母,因其具备高产油脂和脂肪酸的天然能力,因此成为了代谢工程改造微生物生产多不饱和脂肪酸的首选底盘细胞之一。本文首先介绍了多不饱和脂肪酸的来源及其天然合成途径;然后总结归纳了当前利用代谢工程策略改造解脂耶氏酵母生产多不饱和脂肪酸的研究现状;最后对利用解脂耶氏酵母细胞工厂生产多不饱和脂肪酸存在的主要瓶颈问题和未来发展趋势进行了探讨,期望为未来高效合成多不饱和脂肪酸微生物细胞工厂的构建提供理论支持与思路。
何思成, 张紫瑗, 韩雨晴, 苗琳, 张翠英, 于爱群. 解脂耶氏酵母细胞工厂生产多不饱和脂肪酸的研究进展[J]. 生物技术通报, 2024, 40(1): 72-85.
HE Si-cheng, ZHANG Zi-yuan, HAN Yu-qing, MIAO Lin, ZHANG Cui-ying, YU Ai-qun. Research Progress in the Production of Polyunsaturated Fatty Acids by Yarrowia lipolytica Cell Factories[J]. Biotechnology Bulletin, 2024, 40(1): 72-85.
| 名称Name | 学名Scientific name | 简称Abbreviation | 功能Functions |
|---|---|---|---|
| ω-3 PUFA | |||
| α-亚麻酸(α-linolenic acid, ALA) | ∆9, ∆12, ∆15-十八碳三烯酸 | Cl8:3n-3或ω-3 18:3 | 抗血栓、降血脂、预防心肌梗塞、维持视网膜等神经系统所必需的营养因子等 |
| 二十碳五烯酸(Eicosapentaenoic acid, EPA) | ∆5, ∆8, ∆11, ∆14, ∆17-二十碳五烯酸 | C20:5n-3或ω-3 20:5 | 降低心血管疾病的发病率、改善免疫细胞功能、预防及治疗糖尿病等代谢类疾病、预防阿尔兹海默症等 |
| 二十二碳五烯酸(Docosapentaenoic acid, DPA) | ∆7, ∆10, ∆13, ∆16, ∆19-二十二碳五烯酸 | C22:5n-3或ω-3 22:5 | 抑制炎症、抑制血小板凝聚、对颈动脉粥样硬化具有保护作用、抑制神经炎症等 |
| 二十二碳六烯酸(Docosahexaenoic acid, DHA) | ∆4, ∆7, ∆10, ∆13, ∆16, ∆19-二十二碳六烯酸 | C22:6n-3或ω-3 22:6 | 促进脑细胞及神经系统发育、预防心脑血管疾病,调节中枢神经功能等 |
| ω-6 PUFA | |||
| 亚油酸(Linolenic acid, LA) | ∆9, ∆12-十八碳二烯酸 | Cl8:2n-6或ω-6 18:2 | 预防代谢疾病和癌症、抗动脉粥样硬化、抑制肥胖、调节免疫系统等 |
| γ-亚麻酸(γ-linolenic acid, GLA) | ∆6, ∆9, ∆12-十八碳三烯酸 | Cl8:3n-6或ω-6 18:3 | 抗炎症、抗心血管疾病、抗肿瘤、抗糖尿病、缓解更年期综合征等 |
| 二高-γ-亚麻酸(Dohomo-γ-linolenic acid, DGLA) | ∆6, ∆9, ∆12-二十碳三烯酸 | C20:3n-6或ω-6 20:3 | 抑制血栓素活性、抑制血小板凝聚及细胞分裂素分泌、抗动脉粥样硬化等 |
| 花生四烯酸(Arachidonic acid, ARA) | ∆5, ∆8, ∆11, ∆14-二十碳四烯酸 | C20:4n-6或ω-6 20:4 | 合成人体前列腺素的重要前体物质、促进婴幼儿大脑发育、抑制人体各类型皮肤疾病病理发展等 |
表1 生物体中常见的多不饱和脂肪酸类型
Table 1 The most common types of PUFA in organisms
| 名称Name | 学名Scientific name | 简称Abbreviation | 功能Functions |
|---|---|---|---|
| ω-3 PUFA | |||
| α-亚麻酸(α-linolenic acid, ALA) | ∆9, ∆12, ∆15-十八碳三烯酸 | Cl8:3n-3或ω-3 18:3 | 抗血栓、降血脂、预防心肌梗塞、维持视网膜等神经系统所必需的营养因子等 |
| 二十碳五烯酸(Eicosapentaenoic acid, EPA) | ∆5, ∆8, ∆11, ∆14, ∆17-二十碳五烯酸 | C20:5n-3或ω-3 20:5 | 降低心血管疾病的发病率、改善免疫细胞功能、预防及治疗糖尿病等代谢类疾病、预防阿尔兹海默症等 |
| 二十二碳五烯酸(Docosapentaenoic acid, DPA) | ∆7, ∆10, ∆13, ∆16, ∆19-二十二碳五烯酸 | C22:5n-3或ω-3 22:5 | 抑制炎症、抑制血小板凝聚、对颈动脉粥样硬化具有保护作用、抑制神经炎症等 |
| 二十二碳六烯酸(Docosahexaenoic acid, DHA) | ∆4, ∆7, ∆10, ∆13, ∆16, ∆19-二十二碳六烯酸 | C22:6n-3或ω-3 22:6 | 促进脑细胞及神经系统发育、预防心脑血管疾病,调节中枢神经功能等 |
| ω-6 PUFA | |||
| 亚油酸(Linolenic acid, LA) | ∆9, ∆12-十八碳二烯酸 | Cl8:2n-6或ω-6 18:2 | 预防代谢疾病和癌症、抗动脉粥样硬化、抑制肥胖、调节免疫系统等 |
| γ-亚麻酸(γ-linolenic acid, GLA) | ∆6, ∆9, ∆12-十八碳三烯酸 | Cl8:3n-6或ω-6 18:3 | 抗炎症、抗心血管疾病、抗肿瘤、抗糖尿病、缓解更年期综合征等 |
| 二高-γ-亚麻酸(Dohomo-γ-linolenic acid, DGLA) | ∆6, ∆9, ∆12-二十碳三烯酸 | C20:3n-6或ω-6 20:3 | 抑制血栓素活性、抑制血小板凝聚及细胞分裂素分泌、抗动脉粥样硬化等 |
| 花生四烯酸(Arachidonic acid, ARA) | ∆5, ∆8, ∆11, ∆14-二十碳四烯酸 | C20:4n-6或ω-6 20:4 | 合成人体前列腺素的重要前体物质、促进婴幼儿大脑发育、抑制人体各类型皮肤疾病病理发展等 |
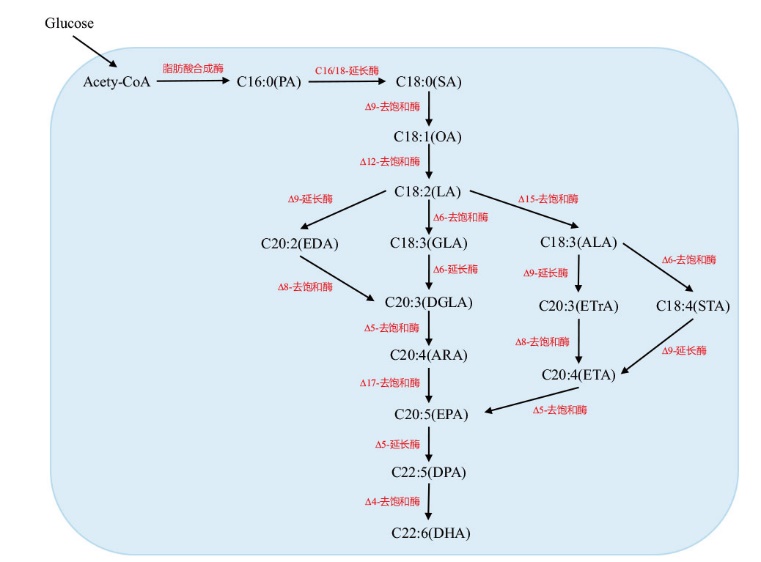
图1 解脂耶氏酵母中PUFA生物合成途径示意图 图中实线表示该酵母细胞中内源存在的PUFA合成途径;虚线表示通过代谢工程在该酵母细胞中所构建的PUFA异源合成途径
Fig. 1 Schematic diagram of the PUFA synthetic pathway in Y. lipolytica In this figure, the solid line indicates the native fatty acid synthesis pathway in Y. lipolytica cells, and the dotted line indicates the heterologous PUFA pathway constructed in Y. lipolytica through metabolic engineering
| 类型Type | 名称Names | 特征Characteristics | 参考文献Reference |
|---|---|---|---|
| 组成型启动子 Constitutive promoters | pTEF | 内源 | [ |
| pTDH1、pGPM1、pFBAIN | 内源 | [ | |
| pEXP1、pGPAT、pGPD | 内源 | [ | |
| pDGA1、pACC、pIDH2、pFAS2、pFAS1、pPOX4、pZWF1、pIDP2 | 内源 | [ | |
| pTEFin | 内源 | [ | |
| 诱导型启动子 Inducible promoters | pGAP、pACL2 | 内源 | [ |
| pXPR2 | 内源;肽诱导型 | [ | |
| pPOX2、pPOT1 | 内源;脂肪酸、烷烃及蓖麻油酸甲酯诱导型;葡萄糖及甘油阻遏型 | [ | |
| pLIP2、pALK1 | 内源;脂肪酸及烷烃诱导型 | [ | |
| pYAT1 | 内源;低氮诱导型 | [ | |
| pEYK1、pEYD1 | 内源;赤藓醇及赤藓糖诱导型;葡萄糖及甘油阻遏型 | [ | |
| pALK1 | 内源;正癸烷诱导型;甘油阻遏型 | [ | |
| pPAT1 | 内源;正癸烷及油酸诱导型;甘油阻遏型 | [ | |
| pTHR1、pMET3、pSER1、pCTR1、pCTR2 | 内源;铜阻遏型 | [ | |
| pHP4d | 杂合 | [ | |
| pnUAS1XPR2-LEU、pnUAS1XPR2-TEF | 杂合 | [ | |
| pEYK1-nAB、pEYK1-4AB-coreTEF, pEYK1/EYD1B-coreEYK1 | 杂合;赤藓醇及赤藓糖诱导型;葡萄糖及甘油阻遏型 | [ | |
| pTEFR1 | 杂合;脂肪酰基辅酶A诱导型 | [ | |
| p(A1R1)×2A3 | 杂合;油酸诱导型 | [ |
表2 解脂耶氏酵母表达系统中常用的启动子
Table 2 Most prominently used promoters for heterologous expression in Y. lipolytica
| 类型Type | 名称Names | 特征Characteristics | 参考文献Reference |
|---|---|---|---|
| 组成型启动子 Constitutive promoters | pTEF | 内源 | [ |
| pTDH1、pGPM1、pFBAIN | 内源 | [ | |
| pEXP1、pGPAT、pGPD | 内源 | [ | |
| pDGA1、pACC、pIDH2、pFAS2、pFAS1、pPOX4、pZWF1、pIDP2 | 内源 | [ | |
| pTEFin | 内源 | [ | |
| 诱导型启动子 Inducible promoters | pGAP、pACL2 | 内源 | [ |
| pXPR2 | 内源;肽诱导型 | [ | |
| pPOX2、pPOT1 | 内源;脂肪酸、烷烃及蓖麻油酸甲酯诱导型;葡萄糖及甘油阻遏型 | [ | |
| pLIP2、pALK1 | 内源;脂肪酸及烷烃诱导型 | [ | |
| pYAT1 | 内源;低氮诱导型 | [ | |
| pEYK1、pEYD1 | 内源;赤藓醇及赤藓糖诱导型;葡萄糖及甘油阻遏型 | [ | |
| pALK1 | 内源;正癸烷诱导型;甘油阻遏型 | [ | |
| pPAT1 | 内源;正癸烷及油酸诱导型;甘油阻遏型 | [ | |
| pTHR1、pMET3、pSER1、pCTR1、pCTR2 | 内源;铜阻遏型 | [ | |
| pHP4d | 杂合 | [ | |
| pnUAS1XPR2-LEU、pnUAS1XPR2-TEF | 杂合 | [ | |
| pEYK1-nAB、pEYK1-4AB-coreTEF, pEYK1/EYD1B-coreEYK1 | 杂合;赤藓醇及赤藓糖诱导型;葡萄糖及甘油阻遏型 | [ | |
| pTEFR1 | 杂合;脂肪酰基辅酶A诱导型 | [ | |
| p(A1R1)×2A3 | 杂合;油酸诱导型 | [ |
| 工程菌株 Engineered strain | 多不饱和脂肪酸 PUFA | 最高产量 Maximum titer | 占总脂肪酸含量Proportion of TFA/% | 参考文献References |
|---|---|---|---|---|
| 小球藻(Chlorella vulgaris) | ALA | — | 10.8 | [ |
| 圆红冬孢酵母(R. kratochvilovae) | ALA | — | 5.9 | [ |
| 解脂耶氏酵母(Y. lipolytica) | ALA | 1 400 mg/L | 28 | [ |
| 裂殖壶菌(Schizochytrium sp.) | EPA | 1 740 mg/L | 1.9 | [ |
| 大肠杆菌(Escherichia coli) | EPA | 31.4 mg/g DCW | — | [ |
| 解脂耶氏酵母(Y. lipolytica) | EPA | 3 200 mg/L | 56.6 | [ |
| 酿酒酵母(S. cerevisiae) | DHA | 39.6 mg/L | 10.2 | [ |
| 恶臭假单孢菌(Paseudomonas putida) | DHA | 1.4 mg/g | — | [ |
| 解脂耶氏酵母(Y. lipolytica) | DHA | 86.5 mg/L(12.8 mg/g DCW) | 10.5 | [ |
| 解脂耶氏酵母(Y. lipolytica) | CLA | 4 000 mg/L | 44 | [ |
| 解脂耶氏酵母(Y. lipolytica) | ARA | 118.1 mg/L | — | [ |
表3 代谢工程改造微生物底盘生产部分多不饱和脂肪酸的产量情况
Table 3 Representative examples of PUFA production by metabolically engineered microorganisms
| 工程菌株 Engineered strain | 多不饱和脂肪酸 PUFA | 最高产量 Maximum titer | 占总脂肪酸含量Proportion of TFA/% | 参考文献References |
|---|---|---|---|---|
| 小球藻(Chlorella vulgaris) | ALA | — | 10.8 | [ |
| 圆红冬孢酵母(R. kratochvilovae) | ALA | — | 5.9 | [ |
| 解脂耶氏酵母(Y. lipolytica) | ALA | 1 400 mg/L | 28 | [ |
| 裂殖壶菌(Schizochytrium sp.) | EPA | 1 740 mg/L | 1.9 | [ |
| 大肠杆菌(Escherichia coli) | EPA | 31.4 mg/g DCW | — | [ |
| 解脂耶氏酵母(Y. lipolytica) | EPA | 3 200 mg/L | 56.6 | [ |
| 酿酒酵母(S. cerevisiae) | DHA | 39.6 mg/L | 10.2 | [ |
| 恶臭假单孢菌(Paseudomonas putida) | DHA | 1.4 mg/g | — | [ |
| 解脂耶氏酵母(Y. lipolytica) | DHA | 86.5 mg/L(12.8 mg/g DCW) | 10.5 | [ |
| 解脂耶氏酵母(Y. lipolytica) | CLA | 4 000 mg/L | 44 | [ |
| 解脂耶氏酵母(Y. lipolytica) | ARA | 118.1 mg/L | — | [ |
| [1] | 赵成, 陈雪芳, 熊莲, 等. 脂溶性荧光染料测定微生物油脂的研究进展[J]. 新能源进展, 2019, 7(1): 85-92. |
| Zhao C, Chen XF, Xiong L, et al. Research progress of determining microbial lipid by liposoluble fluorescent probe[J]. Adv N Renew Energy, 2019, 7(1): 85-92. | |
| [2] |
Carlson SJ, Fallon EM, Kalish BT, et al. The role of the ω-3 fatty acid DHA in the human life cycle[J]. JPEN J Parenter Enteral Nutr, 2013, 37(1): 15-22.
doi: 10.1177/0148607112467821 pmid: 23192455 |
| [3] |
Lorente-Cebrián S, Costa AGV, Navas-Carretero S, et al. Role of omega-3 fatty acids in obesity, metabolic syndrome, and cardiovascular diseases: a review of the evidence[J]. J Physiol Biochem, 2013, 69(3): 633-651.
doi: 10.1007/s13105-013-0265-4 pmid: 23794360 |
| [4] |
Ajami M, Eghtesadi S, Habibey R, et al. Effect of short and long-term treatment with omega-3 Fatty acids on scopolamine-induced amnesia[J]. Iran J Pharm Res, 2012, 11(2): 533-540.
pmid: 24250476 |
| [5] |
袁姚梦, 邢新会, 张翀. 微生物细胞工厂的设计构建:从诱变育种到全基因组定制化创制[J]. 合成生物学, 2020, 1(6): 656-673.
doi: 10.12211/2096-8208.2020-050 |
| Yuan YM, Xing XH, Zhang C. Progress and prospective of engineering microbial cell factories: from random mutagenesis to customized design in genome scale[J]. Synth Biol J, 2020, 1(6): 656-673. | |
| [6] | 赵禹, 刘士琦, 李建, 等. 解脂耶氏酵母作为微生物细胞工厂的应用研究进展[J]. 食品科学, 2021, 42(19): 388-400. |
| Zhao Y, Liu SQ, Li J, et al. Advances in the application of Yarrowia lipolytica as a microbial cell factory[J]. Food Sci, 2021, 42(19): 388-400. | |
| [7] | 王晖, 薛庆节, 杨媛媛, 等. 解脂耶氏酵母在食品工业中的应用[J]. 食品与发酵工业, 2018, 44(8): 291-297. |
| Wang H, Xue QJ, Yang YY, et al. Application of Yarrowia lipolytica in food industry[J]. Food Ferment Ind, 2018, 44(8): 291-297. | |
| [8] |
郑煜堃, 孙青, 陈振, 等. 微生物细胞工厂生产化学品的研究进展——以几种典型小分子和大分子化学品为例[J]. 化工学报, 2021, 72(12): 6109-6121.
doi: 10.11949/0438-1157.20211285 |
| Zheng YK, Sun Q, Chen Z, et al. Progress for chemicals production via microbial cell factory: selecting several small molecules and macromolecular products as examples[J]. CIESC J, 2021, 72(12): 6109-6121. | |
| [9] |
Rossetti RG, Seiler CM, DeLuca P, et al. Oral administration of unsaturated fatty acids: effects on human peripheral blood T lymphocyte proliferation[J]. J Leukoc Biol, 1997, 62(4): 438-443.
doi: 10.1002/jlb.62.4.438 URL |
| [10] |
Chang CS, Sun HL, Lii CK, et al. Gamma-linolenic acid inhibits inflammatory responses by regulating NF-kappaB and AP-1 activation in lipopolysaccharide-induced RAW 264.7 macrophages[J]. Inflammation, 2010, 33(1): 46-57.
doi: 10.1007/s10753-009-9157-8 URL |
| [11] |
Engler MM, Schambelan M, Engler MB, et al. Effects of dietary gamma-linolenic acid on blood pressure and adrenal angiotensin receptors in hypertensive rats[J]. Proc Soc Exp Biol Med, 1998, 218(3): 234-237.
doi: 10.3181/00379727-218-44292 pmid: 9648942 |
| [12] |
Simopoulos AP. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases[J]. Exp Biol Med, 2008, 233(6): 674-688.
doi: 10.3181/0711-MR-311 pmid: 18408140 |
| [13] |
Tumoricidal and anti-angiogenic actions of gamma-linolenic acid and its derivatives[J]. Curr Pharm Biotechnol, 2006, 7(6): 457-466.
doi: 10.2174/138920106779116892 URL |
| [14] |
Escrich E, Moral R, Grau L, et al. Molecular mechanisms of the effects of olive oil and other dietary lipids on cancer[J]. Mol Nutr Food Res, 2007, 51(10): 1279-1292.
pmid: 17879998 |
| [15] |
Tilvis RS, Helve E, Miettinen TA. Improvement of diabetic control by continuous subcutaneous insulin infusion therapy changes fatty acid composition of serum lipids and erythrocytes in type 1(insulin-dependent)diabetes[J]. Diabetologia, 1986, 29(10): 690-694.
pmid: 3803743 |
| [16] |
Hussein N, Ah-Sing E, Wilkinson P, et al. Long-chain conversion of[13C]linoleic acid and alpha-linolenic acid in response to marked changes in their dietary intake in men[J]. J Lipid Res, 2005, 46(2): 269-280.
doi: 10.1194/jlr.M400225-JLR200 pmid: 15576848 |
| [17] |
Tai EKK, Wang XB, Chen ZY. An update on adding docosahexaenoic acid(DHA)and arachidonic acid(AA)to baby formula[J]. Food Funct, 2013, 4(12): 1767-1775.
doi: 10.1039/c3fo60298b URL |
| [18] |
Russell FD, Bürgin-Maunder CS. Distinguishing health benefits of eicosapentaenoic and docosahexaenoic acids[J]. Mar Drugs, 2012, 10(11): 2535-2559.
doi: 10.3390/md10112535 pmid: 23203276 |
| [19] |
Gupta A, Barrow CJ, Puri M. Omega-3 biotechnology: Thraustochytrids as a novel source of omega-3 oils[J]. Biotechnol Adv, 2012, 30(6): 1733-1745.
doi: 10.1016/j.biotechadv.2012.02.014 pmid: 22406165 |
| [20] |
Aasen IM, Ertesvåg H, Heggeset TMB, et al. Thraustochytrids as production organisms for docosahexaenoic acid(DHA), squalene, and carotenoids[J]. Appl Microbiol Biotechnol, 2016, 100(10): 4309-4321.
doi: 10.1007/s00253-016-7498-4 URL |
| [21] |
Ji XJ, Ren LJ, Nie ZK, et al. Fungal arachidonic acid-rich oil: research, development and industrialization[J]. Crit Rev Biotechnol, 2014, 34(3): 197-214.
doi: 10.3109/07388551.2013.778229 URL |
| [22] |
Chang LL, Lu HQ, Chen HQ, et al. Lipid metabolism research in oleaginous fungus Mortierella alpina: current progress and future prospects[J]. Biotechnol Adv, 2022, 54: 107794.
doi: 10.1016/j.biotechadv.2021.107794 URL |
| [23] |
Uemura H. Synthesis and production of unsaturated and polyunsaturated fatty acids in yeast: current state and perspectives[J]. Appl Microbiol Biotechnol, 2012, 95(1): 1-12.
doi: 10.1007/s00253-012-4105-1 pmid: 22562166 |
| [24] |
Xue ZX, Sharpe PL, Hong SP, et al. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica[J]. Nat Biotechnol, 2013, 31(8): 734-740.
doi: 10.1038/nbt.2622 |
| [25] | Zhang HY, Zhang LN, Chen HQ, et al. Enhanced lipid accumulation in the yeast Yarrowia lipolytica by over-expression of ATP: citrate lyase from Mus musculus[J]. J Biotechnol, 2014, 192 Pt A: 78-84. |
| [26] |
Gong YM, Wan X, Jiang ML, et al. Metabolic engineering of microorganisms to produce omega-3 very long-chain polyunsaturated fatty acids[J]. Prog Lipid Res, 2014, 56: 19-35.
doi: 10.1016/j.plipres.2014.07.001 pmid: 25107699 |
| [27] |
Xiong XC, Chen SL. Expanding toolbox for genes expression of Yarrowia lipolytica to include novel inducible, repressible, and hybrid promoters[J]. ACS Synth Biol, 2020, 9(8): 2208-2213.
doi: 10.1021/acssynbio.0c00243 URL |
| [28] | 赵禹, 赵雅坤, 刘士琦, 等. 非常规酵母的分子遗传学及合成生物学研究进展[J]. 微生物学报, 2020, 60(8): 1574-1591. |
| Zhao Y, Zhao YK, Liu SQ, et al. Advances in molecular genetics and synthetic biology tools in unconventional yeasts[J]. Acta Microbiol Sin, 2020, 60(8): 1574-1591. | |
| [29] |
Ogrydziak DM, Scharf SJ. Alkaline extracellular protease produced by Saccharomycopsis lipolytica CX161-1B[J]. J Gen Microbiol, 1982, 128(6): 1225-1234.
pmid: 6750031 |
| [30] |
Müller S, Sandal T, Kamp-Hansen P, et al. Comparison of expression systems in the yeasts Saccharomyces cerevisiae, Hansenula polymorpha, Klyveromyces lactis, Schizosaccharomyces pombe and Yarrowia lipolytica. Cloning of two novel promoters from Yarrowia lipolytica[J]. Yeast, 1998, 14(14): 1267-1283.
pmid: 9802206 |
| [31] |
Larroude M, Rossignol T, Nicaud JM, et al. Synthetic biology tools for engineering Yarrowia lipolytica[J]. Biotechnol Adv, 2018, 36(8): 2150-2164.
doi: S0734-9750(18)30167-8 pmid: 30315870 |
| [32] |
Madzak C, Gaillardin C, Beckerich JM. Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: a review[J]. J Biotechnol, 2004, 109(1-2): 63-81.
doi: 10.1016/j.jbiotec.2003.10.027 URL |
| [33] |
Hong SP, Seip J, Walters-Pollak D, et al. Engineering Yarrowia lipolytica to express secretory invertase with strong FBA1IN promoter[J]. Yeast, 2012, 29(2): 59-72.
doi: 10.1002/yea.v29.2 URL |
| [34] |
Jin EQ, Wong L, Jiao Y, et al. Rapid evolution of regulatory element libraries for tunable transcriptional and translational control of gene expression[J]. Synth Syst Biotechnol, 2017, 2(4): 295-301.
doi: 10.1016/j.synbio.2017.10.003 pmid: 29552654 |
| [35] |
Dulermo R, Brunel F, Dulermo T, et al. Using a vector pool containing variable-strength promoters to optimize protein production in Yarrowia lipolytica[J]. Microb Cell Fact, 2017, 16(1): 31.
doi: 10.1186/s12934-017-0647-3 pmid: 28212656 |
| [36] | Madzak C, Tréton B, Blanchin-Roland S. Strong hybrid promoters and integrative expression/secretion vectors for quasi-constitutive expression of heterologous proteins in the yeast Yarrowia lipolytica[J]. J Mol Microbiol Biotechnol, 2000, 2(2): 207-216. |
| [37] |
Blazeck J, Liu LQ, Redden H, et al. Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach[J]. Appl Environ Microbiol, 2011, 77(22): 7905-7914.
doi: 10.1128/AEM.05763-11 URL |
| [38] |
Blazeck J, Reed B, Garg R, et al. Generalizing a hybrid synthetic promoter approach in Yarrowia lipolytica[J]. Appl Microbiol Biotechnol, 2013, 97(7): 3037-3052.
doi: 10.1007/s00253-012-4421-5 pmid: 23053080 |
| [39] |
Shabbir Hussain M, Gambill L, Smith S, et al. Engineering promoter architecture in oleaginous yeast Yarrowia lipolytica[J]. ACS Synth Biol, 2016, 5(3): 213-223.
doi: 10.1021/acssynbio.5b00100 URL |
| [40] |
Lv YK, Gu Y, Xu JL, et al. Coupling metabolic addiction with negative autoregulation to improve strain stability and pathway yield[J]. Metab Eng, 2020, 61: 79-88.
doi: S1096-7176(20)30093-8 pmid: 32445959 |
| [41] |
Tai M, Stephanopoulos G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production[J]. Metab Eng, 2013, 15: 1-9.
doi: 10.1016/j.ymben.2012.08.007 URL |
| [42] |
Juretzek T, Wang HJ, Nicaud JM, et al. Comparison of promoters suitable for regulated overexpression of β-galactosidase in the alkane-utilizing yeastYarrowia lipolytica[J]. Biotechnol Bioprocess Eng, 2000, 5(5): 320-326.
doi: 10.1007/BF02942206 URL |
| [43] |
Sassi H, Delvigne F, Kar T, et al. Deciphering how LIP2 and POX2 promoters can optimally regulate recombinant protein production in the yeast Yarrowia lipolytica[J]. Microb Cell Fact, 2016, 15(1): 159.
doi: 10.1186/s12934-016-0558-8 URL |
| [44] |
Trassaert M, Vandermies M, Carly F, et al. New inducible promoter for gene expression and synthetic biology in Yarrowia lipolytica[J]. Microb Cell Fact, 2017, 16(1): 141.
doi: 10.1186/s12934-017-0755-0 pmid: 28810867 |
| [45] |
Mori K, Iwama R, Kobayashi S, et al. Transcriptional repression by glycerol of genes involved in the assimilation of n-alkanes and fatty acids in yeast Yarrowia lipolytica[J]. FEMS Yeast Res, 2013, 13(2): 233-240.
doi: 10.1111/fyr.2013.13.issue-2 URL |
| [46] | Park YK, Korpys P, Kubiak M, et al. Engineering the architecture of erythritol-inducible promoters for regulated and enhanced gene expression in Yarrowia lipolytica[J]. FEMS Yeast Res, 2019, 19(1): 10.1093/femsyr/foy105. |
| [47] |
Park BG, Kim J, Kim EJ, et al. Application of random mutagenesis and synthetic FadR promoter for de novo production of ω-hydroxy fatty acid in Yarrowia lipolytica[J]. Front Bioeng Biotechnol, 2021, 9: 624838.
doi: 10.3389/fbioe.2021.624838 URL |
| [48] |
Larroude M, Celinska E, Back A, et al. A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of β-carotene[J]. Biotechnol Bioeng, 2018, 115(2): 464-472.
doi: 10.1002/bit.v115.2 URL |
| [49] |
Gao SL, Han LN, Zhu L, et al. One-step integration of multiple genes into the oleaginous yeast Yarrowia lipolytica[J]. Biotechnol Lett, 2014, 36(12): 2523-2528.
doi: 10.1007/s10529-014-1634-y URL |
| [50] | Wong L, Holdridge B, Engel J, et al. Genetic tools for streamlined and accelerated pathway engineering in Yarrowia lipolytica[J]. Methods Mol Biol, 2019, 1927: 155-177. |
| [51] |
Gibson DG, Young L, Chuang RY, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases[J]. Nat Methods, 2009, 6(5): 343-345.
doi: 10.1038/nmeth.1318 pmid: 19363495 |
| [52] |
Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability[J]. PLoS One, 2008, 3(11): e3647.
doi: 10.1371/journal.pone.0003647 URL |
| [53] |
Ma JB, Gu Y, Marsafari M, et al. Synthetic biology, systems biology, and metabolic engineering of Yarrowia lipolytica toward a sustainable biorefinery platform[J]. J Ind Microbiol Biotechnol, 2020, 47(9-10): 845-862.
doi: 10.1007/s10295-020-02290-8 URL |
| [54] |
Ding Y, Wang KF, Wang WJ, et al. Increasing the homologous recombination efficiency of eukaryotic microorganisms for enhanced genome engineering[J]. Appl Microbiol Biotechnol, 2019, 103(11): 4313-4324.
doi: 10.1007/s00253-019-09802-2 pmid: 31016357 |
| [55] |
Cui ZY, Zheng HH, Jiang ZN, et al. Identification and characterization of the mitochondrial replication origin for stable and episomal expression in Yarrowia lipolytica[J]. ACS Synth Biol, 2021, 10(4): 826-835.
doi: 10.1021/acssynbio.0c00619 URL |
| [56] |
Ji QC, Mai J, Ding Y, et al. Improving the homologous recombination efficiency of Yarrowia lipolytica by grafting heterologous component from Saccharomyces cerevisiae[J]. Metab Eng Commun, 2020, 11: e00152.
doi: 10.1016/j.mec.2020.e00152 URL |
| [57] |
Bai QY, Cheng S, Zhang JL, et al. Establishment of genomic library technology mediated by non-homologous end joining mechanism in Yarrowia lipolytica[J]. Sci China Life Sci, 2021, 64(12): 2114-2128.
doi: 10.1007/s11427-020-1885-x |
| [58] |
Jang IS, Yu BJ, Jang JY, et al. Improving the efficiency of homologous recombination by chemical and biological approaches in Yarrowia lipolytica[J]. PLoS One, 2018, 13(3): e0194954.
doi: 10.1371/journal.pone.0194954 URL |
| [59] | Wagner JM, Williams EV, Alper HS. Developing a piggyBac transposon system and compatible selection markers for insertional mutagenesis and genome engineering in Yarrowia lipolytica[J]. Biotechnol J, 2018, 13(5): e1800022. |
| [60] |
Gao SL, Tong YY, Zhu L, et al. Iterative integration of multiple-copy pathway genes in Yarrowia lipolytica for heterologous β-carotene production[J]. Metab Eng, 2017, 41: 192-201.
doi: 10.1016/j.ymben.2017.04.004 URL |
| [61] |
Lv YK, Edwards H, Zhou JW, et al. Combining 26S rDNA and the cre-loxP system for iterative gene integration and efficient marker curation in Yarrowia lipolytica[J]. ACS Synth Biol, 2019, 8(3): 568-576.
doi: 10.1021/acssynbio.8b00535 URL |
| [62] |
Lv YK, Marsafari M, Koffas M, et al. Optimizing oleaginous yeast cell factories for flavonoids and hydroxylated flavonoids biosynthesis[J]. ACS Synth Biol, 2019, 8(11): 2514-2523.
doi: 10.1021/acssynbio.9b00193 pmid: 31622552 |
| [63] |
Lv YK, Qian S, Du GC, et al. Coupling feedback genetic circuits with growth phenotype for dynamic population control and intelligent bioproduction[J]. Metab Eng, 2019, 54: 109-116.
doi: S1096-7176(19)30028-X pmid: 30940507 |
| [64] |
Zhu HZ, Jiang S, Wu JJ, et al. Production of high levels of 3 S, 3’ S-astaxanthin in Yarrowia lipolytica via iterative metabolic engineering[J]. J Agric Food Chem, 2022, 70(8): 2673-2683.
doi: 10.1021/acs.jafc.1c08072 URL |
| [65] |
Zhou QH, Jiao LC, Li WJ, et al. A novel cre/lox-based genetic tool for repeated, targeted and markerless gene integration in Yarrowia lipolytica[J]. Int J Mol Sci, 2021, 22(19): 10739.
doi: 10.3390/ijms221910739 URL |
| [66] |
Guo ZP, Borsenberger V, Croux C, et al. An artificial chromosome ylAC enables efficient assembly of multiple genes in Yarrowia lipolytica for biomanufacturing[J]. Commun Biol, 2020, 3(1): 199.
doi: 10.1038/s42003-020-0936-y |
| [67] |
DiCarlo JE, Norville JE, Mali P, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems[J]. Nucleic Acids Res, 2013, 41(7): 4336-4343.
doi: 10.1093/nar/gkt135 pmid: 23460208 |
| [68] |
Schwartz CM, Hussain MS, Blenner M, et al. Synthetic RNA polymerase III promoters facilitate high-efficiency CRISPR-Cas9-mediated genome editing in Yarrowia lipolytica[J]. ACS Synth Biol, 2016, 5(4): 356-359.
doi: 10.1021/acssynbio.5b00162 pmid: 26714206 |
| [69] |
Schwartz C, Shabbir-Hussain M, Frogue K, et al. Standardized markerless gene integration for pathway engineering in Yarrowia lipolytica[J]. ACS Synth Biol, 2017, 6(3): 402-409.
doi: 10.1021/acssynbio.6b00285 pmid: 27989123 |
| [70] |
Weninger A, Hatzl AM, Schmid C, et al. Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris[J]. J Biotechnol, 2016, 235: 139-149.
doi: 10.1016/j.jbiotec.2016.03.027 pmid: 27015975 |
| [71] |
Horwitz AA, Walter JM, Schubert MG, et al. Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-cas[J]. Cell Syst, 2015, 1(1): 88-96.
doi: 10.1016/j.cels.2015.02.001 pmid: 27135688 |
| [72] |
Jacobs JZ, Ciccaglione KM, Tournier V, et al. Implementation of the CRISPR-Cas9 system in fission yeast[J]. Nat Commun, 2014, 5: 5344.
doi: 10.1038/ncomms6344 pmid: 25352017 |
| [73] |
Gao SL, Tong YY, Wen ZQ, et al. Multiplex gene editing of the Yarrowia lipolytica genome using the CRISPR-Cas9 system[J]. J Ind Microbiol Biotechnol, 2016, 43(8): 1085-1093.
doi: 10.1007/s10295-016-1789-8 URL |
| [74] | Holkenbrink C, Dam MI, Kildegaard KR, et al. EasyCloneYALI: CRISPR/Cas9-based synthetic toolbox for engineering of the yeast Yarrowia lipolytica[J]. Biotechnol J, 2018, 13(9): e1700543. |
| [75] | Gao DF, Smith S, Spagnuolo M, et al. Dual CRISPR-Cas9 cleavage mediated gene excision and targeted integration in Yarrowia lipolytica[J]. Biotechnol J, 2018, 13(9): e1700590. |
| [76] |
Larroude M, Trabelsi H, Nicaud JM, et al. A set of Yarrowia lipolytica CRISPR/Cas9 vectors for exploiting wild-type strain diversity[J]. Biotechnol Lett, 2020, 42(5): 773-785.
doi: 10.1007/s10529-020-02805-4 |
| [77] |
Schwartz C, Cheng JF, Evans R, et al. Validating genome-wide CRISPR-Cas9 function improves screening in the oleaginous yeast Yarrowia lipolytica[J]. Metab Eng, 2019, 55: 102-110.
doi: S1096-7176(19)30112-0 pmid: 31216436 |
| [78] |
Ng TK, Yu AQ, Ling H, et al. Engineering Yarrowia lipolytica towards food waste bioremediation: production of fatty acid ethyl esters from vegetable cooking oil[J]. J Biosci Bioeng, 2020, 129(1): 31-40.
doi: 10.1016/j.jbiosc.2019.06.009 URL |
| [79] |
Qi BX, Fraser T, Mugford S, et al. Production of very long chain polyunsaturated omega-3 and omega-6 fatty acids in plants[J]. Nat Biotechnol, 2004, 22(6): 739-745.
doi: 10.1038/nbt972 pmid: 15146198 |
| [80] |
Liu HH, Ji XJ, Huang H. Biotechnological applications of Yarrowia lipolytica: past, present and future[J]. Biotechnol Adv, 2015, 33(8): 1522-1546.
doi: 10.1016/j.biotechadv.2015.07.010 pmid: 26248319 |
| [81] |
Dobrowolski A, Drzymała K, Mituła P, et al. Production of tailor-made fatty acids from crude glycerol at low pH by Yarrowia lipolytica[J]. Bioresour Technol, 2020, 314: 123746.
doi: 10.1016/j.biortech.2020.123746 URL |
| [82] |
Christen S, Sauer U. Intracellular characterization of aerobic glucose metabolism in seven yeast species by 13C flux analysis and metabolomics[J]. FEMS Yeast Res, 2011, 11(3): 263-272.
doi: 10.1111/j.1567-1364.2010.00713.x pmid: 21205161 |
| [83] |
Bati N, Hammond EG, Glatz BA. Biomodification of fats and oils: trials withCandida lipolytica[J]. J Am Oil Chem Soc, 1984, 61(11): 1743-1746.
doi: 10.1007/BF02582139 URL |
| [84] |
Park YK, Ledesma-Amaro R. What makes Yarrowia lipolytica well suited for industry?[J]. Trends Biotechnol, 2023, 41(2): 242-254.
doi: 10.1016/j.tibtech.2022.07.006 URL |
| [85] |
Hamilton ML, Powers S, Napier JA, et al. Heterotrophic production of omega-3 long-chain polyunsaturated fatty acids by trophically converted marine diatom Phaeodactylum tricornutum[J]. Mar Drugs, 2016, 14(3): 53.
doi: 10.3390/md14030053 URL |
| [86] |
Damude HG, Zhang HX, Farrall L, et al. Identification of bifunctional delta12/omega3 fatty acid desaturases for improving the ratio of omega3 to omega6 fatty acids in microbes and plants[J]. Proc Natl Acad Sci USA, 2006, 103(25): 9446-9451.
doi: 10.1073/pnas.0511079103 pmid: 16763049 |
| [87] |
Cordova LT, Alper HS. Production of α-linolenic acid in Yarrowia lipolytica using low-temperature fermentation[J]. Appl Microbiol Biotechnol, 2018, 102(20): 8809-8816.
doi: 10.1007/s00253-018-9349-y pmid: 30196328 |
| [88] |
Tezaki S, Iwama R, Kobayashi S, et al. Δ12-fatty acid desaturase is involved in growth at low temperature in yeast Yarrowia lipolytica[J]. Biochem Biophys Res Commun, 2017, 488(1): 165-170.
doi: 10.1016/j.bbrc.2017.05.028 URL |
| [89] | Li HB, Alper HS. Producing biochemicals in Yarrowia lipolytica from xylose through a strain mating approach[J]. Biotechnol J, 2020, 15(2): e1900304. |
| [90] |
Salunke D, Manglekar R, Gadre R, et al. Production of polyunsaturated fatty acids in recombinant Lipomyces starkeyi through submerged fermentation[J]. Bioprocess Biosyst Eng, 2015, 38(7): 1407-1414.
doi: 10.1007/s00449-015-1382-y URL |
| [91] |
Xie DM, Jackson EN, Zhu Q. Sustainable source of omega-3 eicosapentaenoic acid from metabolically engineered Yarrowia lipolytica: from fundamental research to commercial production[J]. Appl Microbiol Biotechnol, 2015, 99(4): 1599-1610.
doi: 10.1007/s00253-014-6318-y URL |
| [92] |
Gu ZN, Shan K, Chen HQ, et al. N-3 polyunsaturated fatty acids and their role in cancer chemoprevention[J]. Curr Pharmacol Rep, 2015, 1(5): 283-294.
doi: 10.1007/s40495-015-0043-9 pmid: 26457243 |
| [93] |
Chen Y, Meesapyodsuk D, Qiu X. Transgenic production of omega-3 very long chain polyunsaturated fatty acids in plants: accomplishment and challenge[J]. Biocatal Agric Biotechnol, 2014, 3(1): 38-43.
doi: 10.1016/j.bcab.2013.08.007 URL |
| [94] |
Gemperlein K, Dietrich D, Kohlstedt M, et al. Polyunsaturated fatty acid production by Yarrowia lipolytica employing designed myxobacterial PUFA synthases[J]. Nat Commun, 2019, 10(1): 4055.
doi: 10.1038/s41467-019-12025-8 pmid: 31492836 |
| [95] |
Sun ML, Madzak C, Liu HH, et al. Engineering Yarrowia lipolytica for efficient γ-linolenic acid production[J]. Biochem Eng J, 2017, 117: 172-180.
doi: 10.1016/j.bej.2016.10.014 URL |
| [96] |
Zhang BX, Rong CC, Chen HQ, et al. De novo synthesis of trans-10, cis-12 conjugated linoleic acid in oleaginous yeast Yarrowia lipolytica[J]. Microb Cell Fact, 2012, 11: 51.
doi: 10.1186/1475-2859-11-51 pmid: 22545818 |
| [97] |
Zhang BX, Chen HQ, Li M, et al. Genetic engineering of Yarrowia lipolytica for enhanced production of trans-10, cis-12 conjugated linoleic acid[J]. Microb Cell Fact, 2013, 12: 70.
doi: 10.1186/1475-2859-12-70 URL |
| [98] |
Liu HH, Madzak C, Sun ML, et al. Engineering Yarrowia lipolytica for arachidonic acid production through rapid assembly of metabolic pathway[J]. Biochem Eng J, 2017, 119: 52-58.
doi: 10.1016/j.bej.2016.12.004 URL |
| [99] |
Norashikin MN, Loh SH, Aziz A, et al. Metabolic engineering of fatty acid biosynthesis in Chlorella vulgaris using an endogenous omega-3 fatty acid desaturase gene with its promoter[J]. Algal Res, 2018, 31: 262-275.
doi: 10.1016/j.algal.2018.02.020 URL |
| [100] | 肖虎. 硫化叶菌病毒基因ORF2促进红冬孢酵母产多不饱和脂肪酸和类胡萝卜素的研究[D]. 昆明: 昆明理工大学, 2018. |
| Xiao H. ORF2 gene of Sulfolobus promoting polyunsaturated fatty acids and carotenoids produced by Rhodosporidium kratochvilovae[D]. Kunming: Kunming University of Science and Technology, 2018. | |
| [101] | 李志朋. 裂殖壶菌利用聚酮合成酶途径合成ω-3多不饱和脂肪酸代谢机制[D]. 厦门: 厦门大学, 2019. |
| Li ZP. Metabolic mechanism of ω-3 polyunsaturated fatty acid synthesis in Schizochytrium through polyketide synthase pathway[D]. Xiamen: Xiamen University, 2019. | |
| [102] |
Amiri-Jami M, Abdelhamid AG, Hazaa M, et al. Recombinant production of omega-3 fatty acids by probiotic Escherichia coli Nissle 1917[J]. FEMS Microbiol Lett, 2015, 362(20): fnv166.
doi: 10.1093/femsle/fnv166 URL |
| [103] | 张明亮. 海洋微生物聚酮合酶途径合成不饱和脂肪酸的分子调控机制研究[D]. 福州: 福建师范大学, 2019. |
| Zhang ML. Molecular regulation mechanism of biosynthesis unsaturated fatty acids by polyketide synthase pathway in marine microbes[D]. Fuzhou: Fujian Normal University, 2019. | |
| [104] |
Gemperlein K, Zipf G, Bernauer HS, et al. Metabolic engineering of Pseudomonas putida for production of docosahexaenoic acid based on a myxobacterial PUFA synthase[J]. Metab Eng, 2016, 33: 98-108.
doi: S1096-7176(15)00150-0 pmid: 26617065 |
| [105] |
Liu HH, Wang C, Lu XY, et al. Improved production of arachidonic acid by combined pathway engineering and synthetic enzyme fusion in Yarrowia lipolytica[J]. J Agric Food Chem, 2019, 67(35): 9851-9857.
doi: 10.1021/acs.jafc.9b03727 URL |
| [1] | 李亮, 徐姗姗, 姜艳军. 生物合成法生产麦角硫因的研究进展[J]. 生物技术通报, 2024, 40(1): 86-99. |
| [2] | 薛宁, 王瑾, 李世新, 刘叶, 程海娇, 张玥, 毛雨丰, 王猛. 多基因同步调控结合高通量筛选构建高产L-苯丙氨酸的谷氨酸棒杆菌工程菌株[J]. 生物技术通报, 2023, 39(9): 268-280. |
| [3] | 程亚楠, 张文聪, 周圆, 孙雪, 李玉, 李庆刚. 乳酸乳球菌生产2'-岩藻糖基乳糖的途径构建及发酵培养基优化[J]. 生物技术通报, 2023, 39(9): 84-96. |
| [4] | 赵思佳, 王晓璐, 孙纪录, 田健, 张杰. 代谢工程改造毕赤酵母生产赤藓糖醇[J]. 生物技术通报, 2023, 39(8): 137-147. |
| [5] | 李雨真, 梅天秀, 李治文, 王淇, 李俊, 邹岳, 赵心清. 红酵母基因组和代谢工程改造研究进展[J]. 生物技术通报, 2023, 39(7): 67-79. |
| [6] | 郁慧丽, 李爱涛. 细胞色素P450酶在香精香料绿色生物合成中的应用[J]. 生物技术通报, 2023, 39(4): 24-37. |
| [7] | 李海宁, 张红兵, 耿革霞, 李冉, 贾振华. 非天然氨基酸的应用及生物合成策略[J]. 生物技术通报, 2023, 39(12): 43-55. |
| [8] | 许瑾, 李涛, 李楚琳, 朱顺妮, 王忠铭, 向文洲. 温度对真眼点藻生长、总脂及二十碳五烯酸(EPA)合成的影响[J]. 生物技术通报, 2022, 38(6): 261-271. |
| [9] | 邱益彬, 马艳琴, 沙媛媛, 朱逸凡, 苏二正, 雷鹏, 李莎, 徐虹. 解淀粉芽孢杆菌分子遗传操作及其应用研究进展[J]. 生物技术通报, 2022, 38(2): 205-217. |
| [10] | 马艳琴, 邱益彬, 李莎, 徐虹. 透明质酸的生物合成及其代谢工程的研究进展[J]. 生物技术通报, 2022, 38(2): 252-262. |
| [11] | 叶敏, 高教琪, 周雍进. 非常规酵母细胞工厂合成天然产物[J]. 生物技术通报, 2021, 37(8): 12-24. |
| [12] | 叶健文, 陈江楠, 张旭, 吴赴清, 陈国强. 动态调控:一种高效的细胞工厂工程化代谢改造策略[J]. 生物技术通报, 2020, 36(6): 1-12. |
| [13] | 陈永灿, 张建志, 司同. 酿酒酵母中基于CRISPR/dCás9的基因转录调控工具的开发与应用[J]. 生物技术通报, 2020, 36(4): 1-12. |
| [14] | 王珂雯 ,尹雪 ,王宇 ,李玉花. 启动子的选择及优化在酿酒酵母代谢工程中的应用[J]. 生物技术通报, 2018, 34(6): 38-47. |
| [15] | 林贝, 李健秀, 刘雪凌. 木质纤维素水解副产物对乙醇发酵的影响及应对措施[J]. 生物技术通报, 2018, 34(3): 23-30. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||