








生物技术通报 ›› 2024, Vol. 40 ›› Issue (5): 112-119.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1223
张娜( ), 刘梦楠, 屈展帆, 崔祎平, 倪嘉瑶, 王华忠(
), 刘梦楠, 屈展帆, 崔祎平, 倪嘉瑶, 王华忠( )
)
收稿日期:2023-12-29
出版日期:2024-05-26
发布日期:2024-03-21
通讯作者:
王华忠,男,博士,教授,研究方向:植物遗传学; E-mail: skywhz@tjnu.edu.cn作者简介:张娜,女,硕士,研究方向:植物遗传学;E-mail: zn1999227@126.com
基金资助:
ZHANG Na( ), LIU Meng-nan, QU Zhan-fan, CUI Yi-ping, NI Jia-yao, WANG Hua-zhong(
), LIU Meng-nan, QU Zhan-fan, CUI Yi-ping, NI Jia-yao, WANG Hua-zhong( )
)
Received:2023-12-29
Published:2024-05-26
Online:2024-03-21
摘要:
【目的】鉴定重要农作物小麦(Triticum aestivum L.)上参与糖酵解的烯醇化酶家族成员ENO2编码基因TaENO2,分析TaENO2的可变翻译特征及产物蛋白的烯醇化酶活性。【方法】采用生物信息学方法鉴定小麦基因组中的TaENO2基因;选择1个TaENO2为代表,以原生质体为受体通过外源表达方法分析该基因的可变翻译产物,通过原核表达和亲和层析过程纯化该基因的重组蛋白产物并测定其烯醇化酶活性。【结果】小麦的六倍体基因组中含有3个部分同源的TaENO2基因,3个基因的产物蛋白含有保守的烯醇化酶活性中心,编码烯醇化酶蛋白序列第94位甲硫氨酸残基(M94)的ATG密码子(ATGM94)是潜在的内部可变翻译起始位点。在原生质体中,外源导入的TaENO2表达出定位于细胞质和细胞核的全长产物烯醇化酶和定位于细胞核的N端截短产物转录抑制因子TaMBP-1两种蛋白,而外源导入的突变ATGM94后的TaENO2基因只表达全长产物烯醇化酶。获得了纯度较高的可溶性重组蛋白GST-TaENO2,该重组蛋白在体外能够催化由2-磷酸-甘油酸(2-PGA)向磷酸烯醇丙酮酸(PEP)转变的烯醇化酶反应。【结论】小麦基因组含有3个烯醇化酶ENO2编码基因TaENO2。在外源表达的情况下,TaENO2表现可变翻译特征,可分别从第一起始密码子和内部ATGM94密码子处起始表达出全长形式的烯醇化酶蛋白和N端截短形式的TaMBP-1蛋白。原核表达的重组蛋白GST-TaENO2具有体外烯醇化酶活性。
张娜, 刘梦楠, 屈展帆, 崔祎平, 倪嘉瑶, 王华忠. 小麦烯醇化酶基因ENO2的可变翻译分析和原核表达[J]. 生物技术通报, 2024, 40(5): 112-119.
ZHANG Na, LIU Meng-nan, QU Zhan-fan, CUI Yi-ping, NI Jia-yao, WANG Hua-zhong. Alternative Translation of Wheat Enolase-encoding Gene ENO2 and Its Prokaryotic Expression[J]. Biotechnology Bulletin, 2024, 40(5): 112-119.
| 引物名称 Prime name | 引物序列 Primer sequence(5'-3') |
|---|---|
| ENO-m-F | AAGTCCGGAGCTAGCTATGGCGGCGACGATCCAGTC |
| ENO-m-R | GCCCTTGCTCACCATGGCGTATGGCTCCACCGGTGCAC |
| Δ93-m-F | AAGTCCGGAGCTAGCTATGGTTCAGCAGCTTGATGGA |
| Δ93-m-R | GCCCTTGCTCACCATGGCGTATGGCTCCACCGGTGCAC |
| M94A-F1 | AAGTCCGGAGCTAGCTATGGCGGCGACGATCCAGTC |
| M94A-R1 | AACAGCAAAGTTGTCGAGCTCAGTTTGAG |
| M94A-F2 | GCTCGACAACTTTGCTGTTCAGCAGCTTGATGGAACCAAG |
| M94A-R2 | GCCCTTGCTCACCATGGCGTATGGCTCCACCGGTGCAC |
| ENO-GST-F | GATCTGGTTCCGCGTGGATCCATGGCGGCGACGATCCAGTC |
| ENO-GST-R | GTCACGATGCGGCCGCTCGAGGTATGGCTCCACCGGTGCAC |
表1 实验所用引物
Table 1 Primers used in this study
| 引物名称 Prime name | 引物序列 Primer sequence(5'-3') |
|---|---|
| ENO-m-F | AAGTCCGGAGCTAGCTATGGCGGCGACGATCCAGTC |
| ENO-m-R | GCCCTTGCTCACCATGGCGTATGGCTCCACCGGTGCAC |
| Δ93-m-F | AAGTCCGGAGCTAGCTATGGTTCAGCAGCTTGATGGA |
| Δ93-m-R | GCCCTTGCTCACCATGGCGTATGGCTCCACCGGTGCAC |
| M94A-F1 | AAGTCCGGAGCTAGCTATGGCGGCGACGATCCAGTC |
| M94A-R1 | AACAGCAAAGTTGTCGAGCTCAGTTTGAG |
| M94A-F2 | GCTCGACAACTTTGCTGTTCAGCAGCTTGATGGAACCAAG |
| M94A-R2 | GCCCTTGCTCACCATGGCGTATGGCTCCACCGGTGCAC |
| ENO-GST-F | GATCTGGTTCCGCGTGGATCCATGGCGGCGACGATCCAGTC |
| ENO-GST-R | GTCACGATGCGGCCGCTCGAGGTATGGCTCCACCGGTGCAC |

图1 小麦和拟南芥烯醇化酶蛋白的多序列比对 TaENO2-5A、TaENO2-5B和TaENO2-5D为3个部分同源的小麦烯醇化酶ENO2蛋白,AtENO1、AtENO2和AtENO3为拟南芥烯醇化酶家族的3个成员。红色实线框示活性中心的负责结合镁离子的丝氨酸残基(S),红色虚线框示ENO2序列中的第二个甲硫氨酸残基(M),编码该残基的ATG密码子是可变翻译内部起始密码子
Fig. 1 Protein sequence alignment of wheat and Arabidopsis enolases TaENO2-5A, TaENO2-5B, and TaENO2-5D are wheat enolases of homeologous ENO2 proteins, and AtENO1, AtENO2, and AtENO3 are Arabidopsis enolase proteins. Solid red box indicates the serine residue(S)responsible for binding of Mg2+ in the catalytic center. Dashed red box indicates the second methionine residue(M)in the ENO2 sequences. The ATG codon encoding this residue is an internal start codon of alternative translation

图2 TaENO2的可变翻译产物分析 A:3种(I、II和III)融合mCherry的TaENO2编码序列结构模式图;B:TaENO2可变翻译产物的亚细胞定位。将图A中所示的不同mCherry融合基因转化到原生质体中进行表达,对转化后24 h的原生质体进行细胞核的Hoechst 33342染色,随后在共聚焦显微镜下观察原生质体中红色荧光的亚细胞分布。图中比例尺为10 μm;C:Western杂交鉴定在原生质体中表达的TaENO2基因的可变翻译产物
Fig. 2 Alternative translation products of TaENO2 A: Schematic representation of the three forms(I, II, and III)of mCherry-fused TaENO2 coding sequences. B: Subcellular localization of the alternative translation products of TaENO2. Different mCherry fusion genes shown in(A)were respectively transformed into protoplasts for expression. The subcellular distribution of red fluorescence and the Hoechst 33342-stained nuclei in protoplasts were observed at 24 h after protoplast transformation. Scale bar indicates 10 μm. C: Western blot analysis on the alternative translation products of TaENO2 expressed in protoplasts
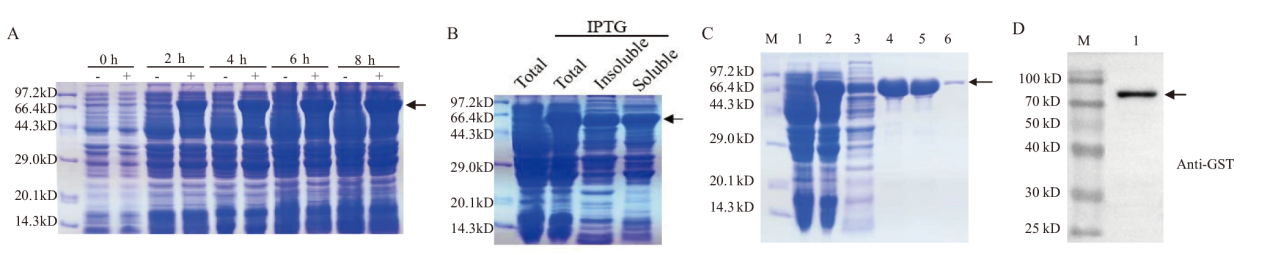
图3 GST-TaENO2重组蛋白的原核表达与纯化 A:GST-TaENO2重组蛋白的原核表达。将携带有GST-TaENO2基因的大肠杆菌BL21(DE3)菌株培养至对数生长期,然后在培养物中加入IPTG(+)诱导重组蛋白的表达至图中所示时间点,同时设置不加IPTG(-)对照培养物。煮沸裂解大肠杆菌培养物释放总蛋白,取等量的总蛋白进行SDS-PAGE分离。箭头示诱导表达的GST-TaENO2重组蛋白条带;B:原核表达重组蛋白的可溶性分析。对经IPTG诱导的大肠杆菌培养物进行超声破碎和可溶、不溶性成分的离心分离,对总蛋白、不溶性蛋白和可溶性蛋白进行电泳分离,箭头示GST-TaENO2重组蛋白条带;C:GST-TaENO2重组蛋白的纯化。M:蛋白分子量标准;1:未经IPTG诱导的大肠杆菌总蛋白;2:经IPTG诱导4 h的大肠杆菌总蛋白;3-6:亲和层析纯化过程中收集的流穿液(3)和连续单管洗脱液(4-6),箭头示重组蛋白条带;D:纯化重组蛋白的Western杂交鉴定
Fig. 3 Prokaryotic expression and purification of recombinant GST-TaENO2 protein A: Prokaryotic expression of recombinant GST-TaENO2 protein. The E. coli strain BL21(DE3)carrying the fusion gene GST-TaENO2 was grown to the log growth phase. The heterologous protein expression was then induced with IPTG for the indicated time periods. Cells were lysed by boiling and an equal aliquot of protein was resolved by SDS-PAGE. Arrows indicate the bands of recombinant protein. B: Solubility analysis on the prokaryotically expressed recombinant GST-TaENO2 protein. E. coli culture induced by IPTG was lysed by sonication. The soluble and insoluble protein fractions were separated by centrifugation and resolved by SDS-PAGE. Arrows indicate the bands of recombinant protein GST-TaENO2. C: Purification of recombinant GST-TaENO2 protein. M: Protein marker; 1: Total extracted protein from the E. coli culture which was not subjected to IPTG induction of gene expression; 2: Total extracted protein from the E.coli culture which had been subjected to IPTG induction of gene expression for 4 h; 3-6: Collected flow-through(3)and successive elution fractions(4-6)in the process of protein purification by affinity chromatography. D: Western blot analysis of the purified recombinant GST-TaENO2 protein. Arrows indicate the bands of recombinant protein
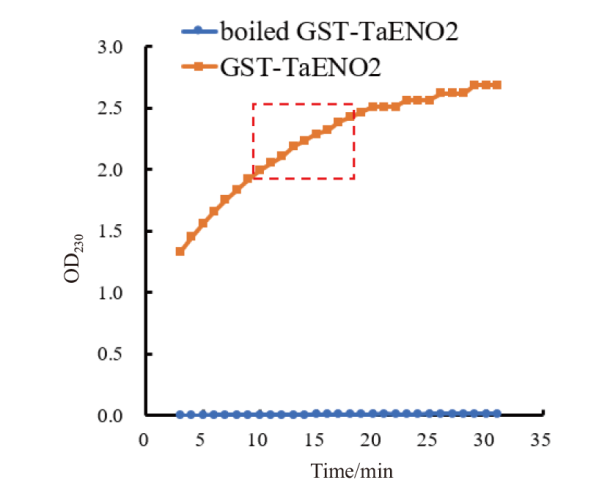
图4 重组蛋白GST-TaENO2的体外烯醇化酶活性分析 烯醇化酶催化产物PEP在230 nm波长处有最大吸收峰。图中所示为体外烯醇化酶活性分析的反应液在230 nm 处的吸光值(OD230)随时间的变化趋势。红色虚线框示OD230的近似线性增长阶段,该阶段的数据用于计算酶活力
Fig. 4 In vitro assays on the enolase activity of the recombinant protein GST-TaENO2 The catalytic product of enolase, PEP, has maximum optical absorption at a wavelength of 230 nm. The figure shows the time-dependent changes in the optical density of the in vitro enolase assay solutions at 230 nm(OD230). Red dash box indicates the near linear increase stage of OD230. The data of this stage were used for calculation of enolase activity
| [1] |
Andriotis VME, Kruger NJ, et al. Plastidial glycolysis in developing Arabidopsis embryos[J]. New Phytol, 2010, 185(3): 649-662.
doi: 10.1111/j.1469-8137.2009.03113.x pmid: 20002588 |
| [2] |
Lee H, Guo Y, Ohta M, et al. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase[J]. EMBO J, 2002, 21(11): 2692-2702.
doi: 10.1093/emboj/21.11.2692 pmid: 12032082 |
| [3] |
Voll LM, Hajirezaei MR, Czogalla-Peter C, et al. Antisense inhibition of enolase strongly limits the metabolism of aromatic amino acids, but has only minor effects on respiration in leaves of transgenic tobacco plants[J]. New Phytol, 2009, 184(3): 607-618.
doi: 10.1111/j.1469-8137.2009.02998.x pmid: 19694966 |
| [4] |
Troncoso-Ponce MA, Rivoal J, Dorion S, et al. Molecular and biochemical characterization of the sunflower(Helianthus annuus L.) cytosolic and plastidial enolases in relation to seed development[J]. Plant Sci, 2018, 272: 117-130.
doi: S0168-9452(18)30173-0 pmid: 29807582 |
| [5] |
Qiao G, Wu AG, Chen XL, et al. Enolase 1, a moonlighting protein, as a potential target for cancer treatment[J]. Int J Biol Sci, 2021, 17(14): 3981-3992.
doi: 10.7150/ijbs.63556 pmid: 34671213 |
| [6] | Liu WC, Song RF, Qiu YM, et al. Sulfenylation of ENOLASE2 facilitates H2O2-conferred freezing tolerance in Arabidopsis[J]. Dev Cell, 2022, 57(15): 1883-1898.e5. |
| [7] | Barkla BJ, Vera-Estrella R, et al. Quantitative proteomics of the tonoplast reveals a role for glycolytic enzymes in salt tolerance[J]. Plant Cell, 2009, 21(12): 4044-4058. |
| [8] | Liu ZJ, Zheng LM, Pu L, et al. ENO2 affects the seed size and weight by adjusting cytokinin content and forming ENO2-bZIP75 complex in Arabidopsis thaliana[J]. Front Plant Sci, 2020, 11: 574316. |
| [9] | Liu ZJ, Liu HM, Zheng LM, et al. Enolase2 regulates seed fatty acid accumulation via mediating carbon partitioning in Arabidopsis thaliana[J]. Physiol Plant, 2022, 174(6): e13797. |
| [10] |
Zhang YJ, Sampathkumar A, Kerber SML, et al. A moonlighting role for enzymes of glycolysis in the co-localization of mitochondria and chloroplasts[J]. Nat Commun, 2020, 11(1): 4509.
doi: 10.1038/s41467-020-18234-w pmid: 32908151 |
| [11] | Eremina M, Rozhon W, et al. ENO2 activity is required for the development and reproductive success of plants, and is feedback-repressed by AtMBP-1[J]. Plant J, 2015, 81(6): 895-906. |
| [12] | Liu ZJ, Zhang YH, Ma XF, et al. Biological functions of Arabidopsis thaliana MBP-1-like protein encoded by ENO2 in the response to drought and salt stresses[J]. Physiol Plant, 2020, 168(3): 660-674. |
| [13] | Yang LY, Wang ZX, Zhang AQ, et al. Reduction of the canonical function of a glycolytic enzyme enolase triggers immune responses that further affect metabolism and growth in Arabidopsis[J]. Plant Cell, 2022, 34(5): 1745-1767. |
| [14] | Ma XF, Wu Y, Ming HN, et al. AtENO2 functions in the development of male gametophytes in Arabidopsis thaliana[J]. J Plant Physiol, 2021, 263: 153417. |
| [15] | Bi CX, Yu YH, Dong CH, et al. The bZIP transcription factor TabZIP15 improves salt stress tolerance in wheat[J]. Plant Biotechnol J, 2021, 19(2): 209-211. |
| [16] | Zhang YF, Wang JY, et al. Wheat TaSnRK2.10 phosphorylates TaERD15 and TaENO1 and confers drought tolerance when overexpressed in rice[J]. Plant Physiol, 2023, 191(2): 1344-1364. |
| [17] | Liu ZJ, Zhang A, Zheng LM, et al. The biological significance and regulatory mechanism of c-myc binding protein 1(MBP-1)[J]. Int J Mol Sci, 2018, 19(12): 3868. |
| [18] | Kang M, Abdelmageed H, Lee S, et al. AtMBP-1, an alternative translation product of LOS2, affects abscisic acid responses and is modulated by the E3 ubiquitin ligase AtSAP5[J]. Plant J, 2013, 76(3): 481-493. |
| [19] |
杜晓敏, 王均, 等. 小麦叶肉细胞原生质体制备参数解析及在基因瞬时表达上的应用[J]. 华北农学报, 2015, 30(6): 52-60.
doi: 10.7668/hbnxb.2015.06.008 |
| Du XM, Wang J, et al. Factors influencing the preparation of wheat mesophyll protoplasts and the application of wheat mesophyll protoplasts in gene transient expression[J]. Acta Agric Boreali Sin, 2015, 30(6): 52-60. |
| [1] | 潘萍萍, 徐志浩, 张怡雯, 李青, 王忠华. 多花黄精查尔酮合酶PcCHS的原核表达、亚细胞定位及表达分析[J]. 生物技术通报, 2024, 40(5): 280-289. |
| [2] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| [3] | 张怡, 张心如, 张金珂, 胡利宗, 上官欣欣, 郑晓红, 胡娟娟, 张聪聪, 穆桂清, 李成伟. 小麦镉胁迫响应基因TaMYB1的功能分析[J]. 生物技术通报, 2024, 40(1): 194-206. |
| [4] | 焦进兰, 王文文, 介欣芮, 王华忠, 岳洁瑜. 外源钙缓解小麦幼苗盐胁迫的作用机制[J]. 生物技术通报, 2024, 40(1): 207-221. |
| [5] | 常泸尹, 王中华, 李凤敏, 高梓源, 张辉红, 王祎, 李芳, 韩燕来, 姜瑛. 玉米根际多功能促生菌的筛选及其对冬小麦-夏玉米轮作体系产量提升效果[J]. 生物技术通报, 2024, 40(1): 231-242. |
| [6] | 温晓蕾, 李建嫄, 李娜, 张娜, 杨文香. 小麦叶锈菌与小麦互作的酵母双杂交cDNA文库构建与应用[J]. 生物技术通报, 2023, 39(9): 136-146. |
| [7] | 韩志阳, 贾子苗, 梁秋菊, 王轲, 唐华丽, 叶兴国, 张双喜. 二套小麦-簇毛麦染色体附加系苗期耐盐性及籽粒硒和叶酸的含量[J]. 生物技术通报, 2023, 39(8): 185-193. |
| [8] | 梅欢, 李玥, 刘可蒙, 刘吉华. 小檗碱桥酶高效原核表达及生物合成l-SLR的研究[J]. 生物技术通报, 2023, 39(7): 277-287. |
| [9] | 滕梦鑫, 徐亚, 何静, 汪奇, 乔飞, 李敬阳, 李新国. 香蕉MaMC6的克隆及原核表达分析[J]. 生物技术通报, 2023, 39(12): 179-186. |
| [10] | 孔德真, 聂迎彬, 崔凤娟, 桑伟, 徐红军, 田笑明. 杂交小麦制种研究现状及展望[J]. 生物技术通报, 2023, 39(1): 95-103. |
| [11] | 郭志浩, 金泽鑫, 刘琦, 高利. 小麦矮腥黑粉菌效应蛋白g11335的生物信息学分析、亚细胞定位及毒性验证[J]. 生物技术通报, 2022, 38(8): 110-117. |
| [12] | 索青青, 吴楠, 杨慧, 李莉, 王锡锋. 水稻咖啡酰辅酶A-O-甲基转移酶基因的原核表达、抗体制备和应用[J]. 生物技术通报, 2022, 38(8): 135-141. |
| [13] | 覃雪晶, 王雨涵, 曹一博, 张凌云. 青杄PwHAP5基因原核表达及多克隆抗体制备[J]. 生物技术通报, 2022, 38(8): 142-149. |
| [14] | 赵静雅, 彭梦雅, 张时雨, 单艺轩, 邢小萍, 施艳, 李海洋, 杨雪, 李洪连, 陈琳琳. C2H2锌指转录因子FpCzf7参与假禾谷镰孢的生长和致病性[J]. 生物技术通报, 2022, 38(8): 216-224. |
| [15] | 王光丽, 范婵, 王辉, 卢惠芳, 夏灵尹, 黄健, 闵迅. 霍乱弧菌溶血素HlyA的原核表达、纯化及多克隆抗体制备与鉴定[J]. 生物技术通报, 2022, 38(7): 269-277. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||