








生物技术通报 ›› 2024, Vol. 40 ›› Issue (5): 248-260.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1174
孙亚楠( ), 王春雪, 王欣, 杜秉海, 刘凯(
), 王春雪, 王欣, 杜秉海, 刘凯( ), 汪城墙(
), 汪城墙( )
)
收稿日期:2023-12-13
出版日期:2024-05-26
发布日期:2024-03-21
通讯作者:
汪城墙,男,博士,副教授,研究方向:微生物代谢与分子生物学 ;E-mail: wangcq@sdau.edu.cn;作者简介:孙亚楠,女,硕士研究生,研究方向:微生物资源挖掘与应用;E-mail: sun15383646425@163.com
基金资助:
SUN Ya-nan( ), WANG Chun-xue, WANG Xin, DU Bing-hai, LIU Kai(
), WANG Chun-xue, WANG Xin, DU Bing-hai, LIU Kai( ), WANG Cheng-qiang(
), WANG Cheng-qiang( )
)
Received:2023-12-13
Published:2024-05-26
Online:2024-03-21
摘要:
【目的】从黄河三角洲盐碱地中筛选到具有抗盐促生作用的一株根际菌,制成菌剂用于缓解玉米植株的高盐胁迫。【方法】通过功能培养基、16S rDNA序列和全基因组分析,确定菌株种属以及对玉米植株的抗盐促生效果。【结果】从黄河三角洲盐碱地中筛选到一株可在0-16%的NaCl和pH 5-8的条件下正常生长的萎缩芽孢杆菌CNY01,该菌株可促进玉米在高盐条件下生长,具有降蛋白和解钾的能力,还可抑制青枯雷尔氏菌、串珠镰刀菌等病原菌的生长。玉米植株在含100 mmol/L NaCl的营养液中生长8 d时,施加菌株CNY01可使植株的生理株高、根长和鲜重分别提高38.80%(P<0.01)、23.73%和28.19%(P<0.01)。玉米植株在加100 mmol/L NaCl的高盐盆栽条件下生长28 d时,菌株CNY01可使植株的株高、地上鲜重和地下鲜重分别提高10.84%、41.87%(P<0.01)和23.29%。全基因组序列分析也预测出该菌株基因组中含有维持细胞渗透压、合成应激蛋白等缓解高盐胁迫的基因。【结论】从黄河三角洲盐碱地中筛选到一株具有防病促生功能的萎缩芽孢杆菌CNY01,对玉米植株有显著的抗盐促生作用,结合基因组结果预测到该菌株含有与抗盐促生相关基因,是重要的菌种资源。
孙亚楠, 王春雪, 王欣, 杜秉海, 刘凯, 汪城墙. 萎缩芽孢杆菌CNY01的生防特性及其对玉米的抗盐促生作用[J]. 生物技术通报, 2024, 40(5): 248-260.
SUN Ya-nan, WANG Chun-xue, WANG Xin, DU Bing-hai, LIU Kai, WANG Cheng-qiang. Biocontrol Characteristics of Bacillus atrophaeus CNY01 and Its Salt-resistant and Growth-promoting Effect on Maize Seedling[J]. Biotechnology Bulletin, 2024, 40(5): 248-260.
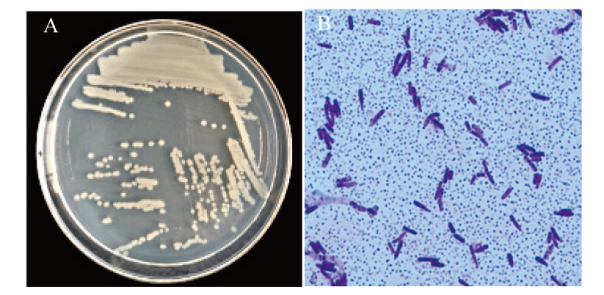
图1 菌株CNY01的菌落和细胞形态特征 A:菌株CNY01菌落图片;B:菌株CNY01的革兰氏染色图片
Fig. 1 Colony and cell morphological characteristics of strain CNY01 A: Colony picture of strain CNY01; B: Gram stain picture of strain CNY01

图3 菌株CNY01的耐盐和耐酸碱能力测定 A:菌株CNY01在不同盐浓度固体培养基中的长势;B:菌株CNY01的液体培养耐盐测定;C:CNY01的液体培养耐酸碱测定
Fig. 3 Determination of salt and acid-alkali tolerance of strain CNY01 A: Growth of CNY01 on solid medium with different salt concentrations; B: salt tolerance test of CNY01 in liquid medium; C: determination of acid and alkali resistance of CNY01 in liquid medium
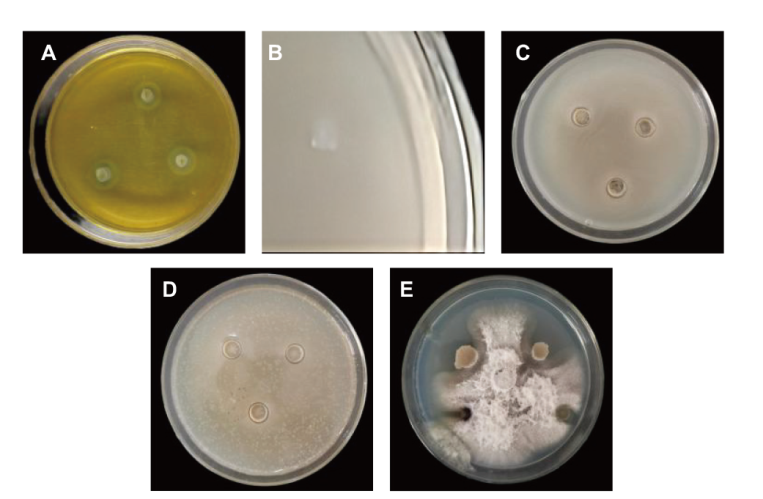
图4 菌株CNY01的拮抗促生能力测定 A:干酪素培养基培养CNY01;B:硅酸盐培养基培养CNY01;C:CNY01与枯草芽孢杆菌拮抗培养;D:CNY01与青枯雷尔氏菌拮抗培养;E:CNY01与串珠镰刀菌拮抗培养
Fig. 4 Determination of antagonistic and growth-promoting ability of strain CNY01 A: CNY01 cultured in casein medium; B: CNY01 cultured in silicate medium; C: CNY01 cultured in antagonism with Bacillus subtilis; D: CNY01 cultured in antagonism with Ralstonia solanacearum; E: CNY01 cultured in antagonism with Fusarium moniliforme
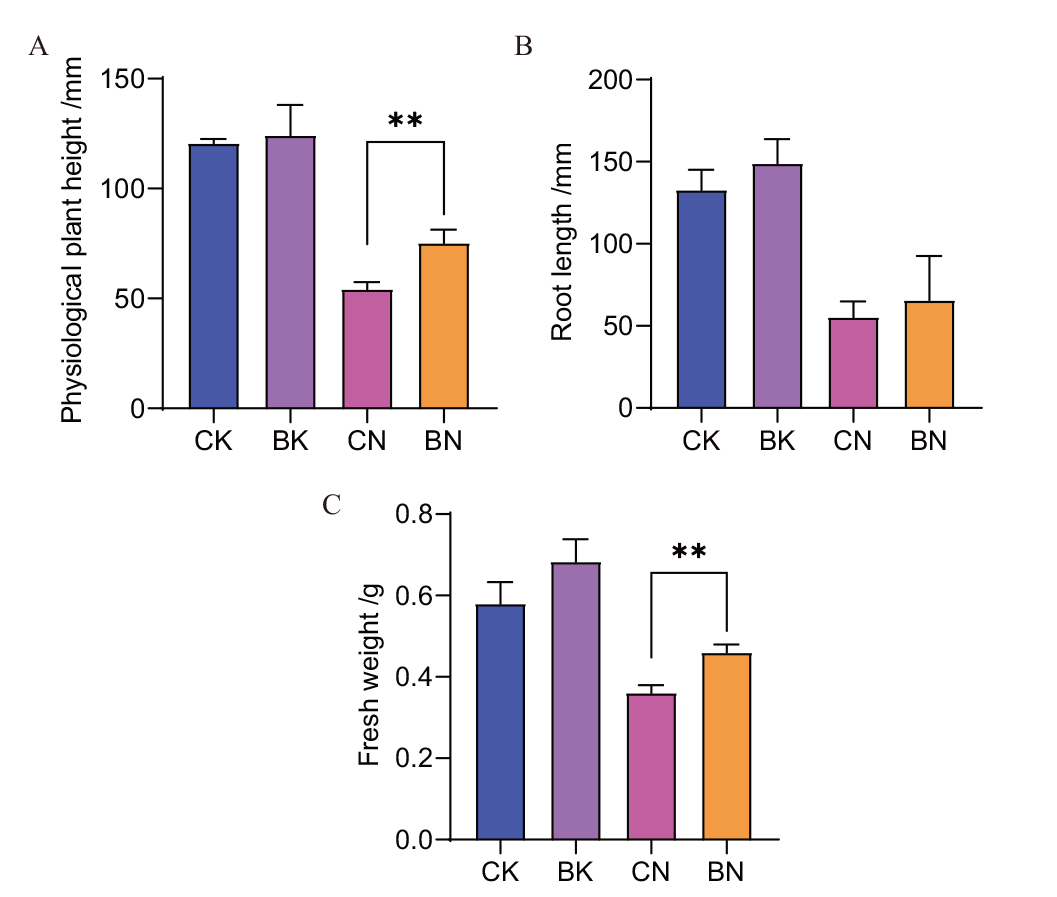
图5 菌株CNY01对玉米植株的促生作用(第8天) A:玉米生理株高;B:玉米根长;C:植株鲜重。CK:无菌无盐组;BK:有菌无盐组;CN:无菌有盐组;BN:有菌有盐组。*P<0.05,**P<0.01,下同
Fig. 5 Growth-promoting effect of strain CNY01 on maize seedlings(Day 8) A: Maize physiological plant height; B: maize root length; C: plant fresh weight. CK: Sterile and salt-free group; BK: the salt-free group with strain CNY01; CN: sterile salt group; BN: the salt group with strain CNY01. *P< 0.05 and **P < 0.01, the same below
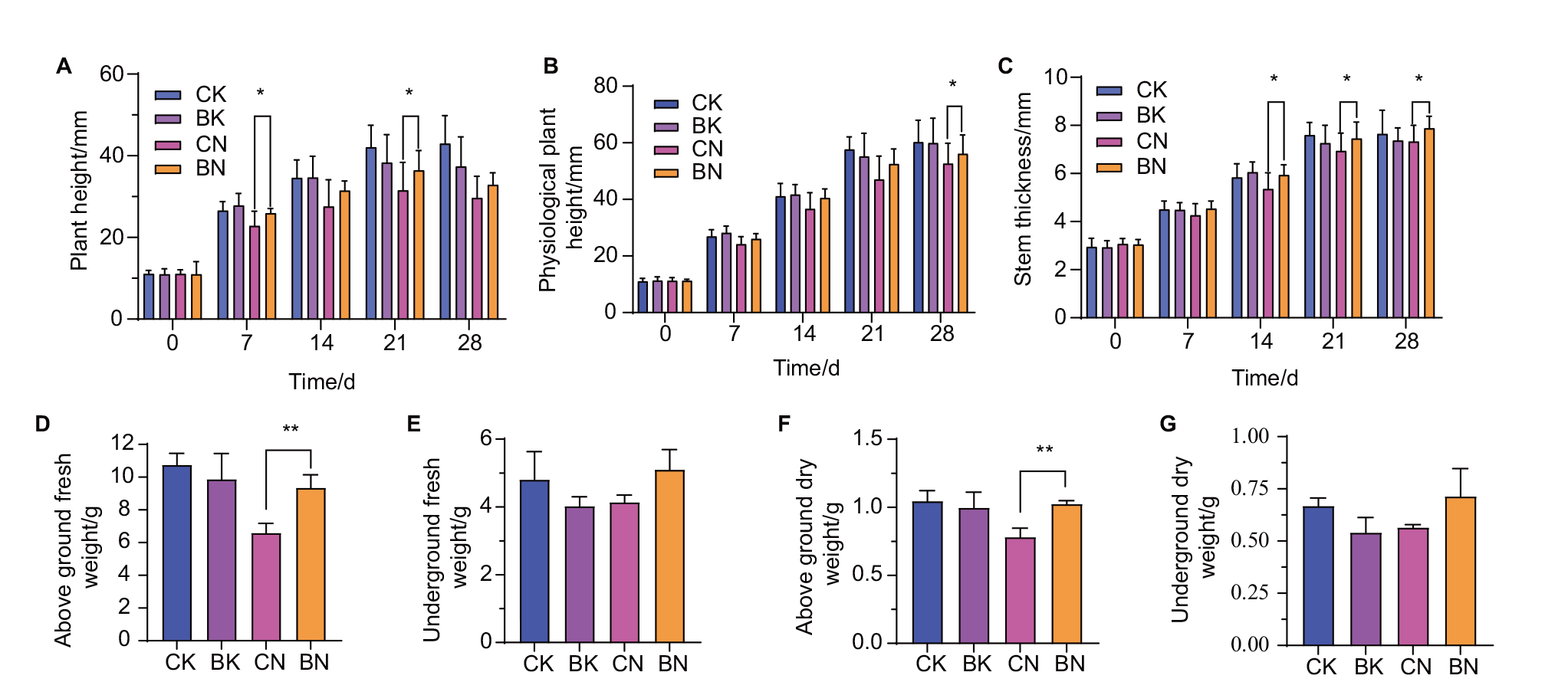
图6 盆栽条件下不同时期玉米植株的农艺性状分析 A:株高;B:生理株高;C:茎粗;D:地上鲜重(28 d);E:地下鲜重(28 d);F:地上干重(28 d);G:地下干重(28 d)
Fig. 6 Analysis of agronomic traits of maize seedlings in different periods under potted conditions A: Plant height; B: physiological plant height; C: stem thick; D: aboveground fresh weight(28 d); E: underground fresh weight(28 d); F: aboveground dry weight(28 d);G: underground dry weight(28 d)
| Genomic information | CNY01 | GQJK17 | BA59 | PENSV20 | 1942 | SRCM101359 | NS2 | NX-12 | MBLB1156 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Genome Size/Mb | 4.14 | 4.33 | 4.26 | 4.15 | 4.17 | 4.18 | 4.26 | 4.16 | 4.15 | |
| G+C content/% | 43.50 | 43.30 | 43.10 | 43.50 | 43.20 | 43.30 | 43.30 | 43.30 | 43.40 | |
| CDSs(with protein) | 3950 | 4045 | 3889 | 3932 | 3995 | 3923 | 4018 | 3946 | 3870 | |
| Pseudo genes number | 146 | 161 | 383 | 157 | 152 | 157 | 159 | 158 | 232 | |
| rRNA genes number | 24 | 24 | 26 | 24 | 21 | 24 | 24 | 24 | 24 | |
| tRNA genes number | 82 | 84 | 84 | 83 | 75 | 82 | 82 | 82 | 82 | |
| The source of isolation | Soil (China) | Rhizophere (China) | Cotton root (China) | Soil (Canada) | - | Food (South Korea) | - | Cotton rhizosphere soil | Fermented to fu |
表1 萎缩芽孢杆菌CNY01、GQJK17、BA59、PENSV20、1942、SRCM101359、NS2、NX-12和MBLB1156基因组信息比对
Table 1 General genome features of B. atrophaeus CNY01, GQJK17, BA59, PENSV20, 1942, SRCM101359, NS2, NX-12, and MBLB1156
| Genomic information | CNY01 | GQJK17 | BA59 | PENSV20 | 1942 | SRCM101359 | NS2 | NX-12 | MBLB1156 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Genome Size/Mb | 4.14 | 4.33 | 4.26 | 4.15 | 4.17 | 4.18 | 4.26 | 4.16 | 4.15 | |
| G+C content/% | 43.50 | 43.30 | 43.10 | 43.50 | 43.20 | 43.30 | 43.30 | 43.30 | 43.40 | |
| CDSs(with protein) | 3950 | 4045 | 3889 | 3932 | 3995 | 3923 | 4018 | 3946 | 3870 | |
| Pseudo genes number | 146 | 161 | 383 | 157 | 152 | 157 | 159 | 158 | 232 | |
| rRNA genes number | 24 | 24 | 26 | 24 | 21 | 24 | 24 | 24 | 24 | |
| tRNA genes number | 82 | 84 | 84 | 83 | 75 | 82 | 82 | 82 | 82 | |
| The source of isolation | Soil (China) | Rhizophere (China) | Cotton root (China) | Soil (Canada) | - | Food (South Korea) | - | Cotton rhizosphere soil | Fermented to fu |
| Name of strain | Bacillus haynesii | Bacillus sonorensis | Bacillus siamensis | Bacillus licheniformis | Bacillus atrophaeus | Bacillus paralicheniformis | CNY01 |
|---|---|---|---|---|---|---|---|
| Bacillus haynesii | * | 81.18 | 71.86 | 95.14 | 72.29 | 95.14 | 72.23 |
| Bacillus sonorensis | 80.85 | * | 71.84 | 80.97 | 72.47 | 80.72 | 72.38 |
| Bacillus siamensis | 72.04 | 71.92 | * | 71.93 | 76.64 | 71.84 | 76.73 |
| Bacillus licheniformis | 95.40 | 81.42 | 71.94 | * | 72.45 | 94.38 | 72.43 |
| Bacillus atrophaeus | 72.28 | 72.57 | 76.66 | 72.30 | * | 72.32 | 98.06 |
| Bacillus paralicheniformis | 95.03 | 81.00 | 71.98 | 94.02 | 72.25 | * | 72.13 |
| CNY01 | 72.32 | 72.63 | 76.89 | 72.33 | 98.36 | 72.34 | * |
表2 菌株CNY01的ANI比对结果
Table 2 ANI contrast test of strain CNY01
| Name of strain | Bacillus haynesii | Bacillus sonorensis | Bacillus siamensis | Bacillus licheniformis | Bacillus atrophaeus | Bacillus paralicheniformis | CNY01 |
|---|---|---|---|---|---|---|---|
| Bacillus haynesii | * | 81.18 | 71.86 | 95.14 | 72.29 | 95.14 | 72.23 |
| Bacillus sonorensis | 80.85 | * | 71.84 | 80.97 | 72.47 | 80.72 | 72.38 |
| Bacillus siamensis | 72.04 | 71.92 | * | 71.93 | 76.64 | 71.84 | 76.73 |
| Bacillus licheniformis | 95.40 | 81.42 | 71.94 | * | 72.45 | 94.38 | 72.43 |
| Bacillus atrophaeus | 72.28 | 72.57 | 76.66 | 72.30 | * | 72.32 | 98.06 |
| Bacillus paralicheniformis | 95.03 | 81.00 | 71.98 | 94.02 | 72.25 | * | 72.13 |
| CNY01 | 72.32 | 72.63 | 76.89 | 72.33 | 98.36 | 72.34 | * |
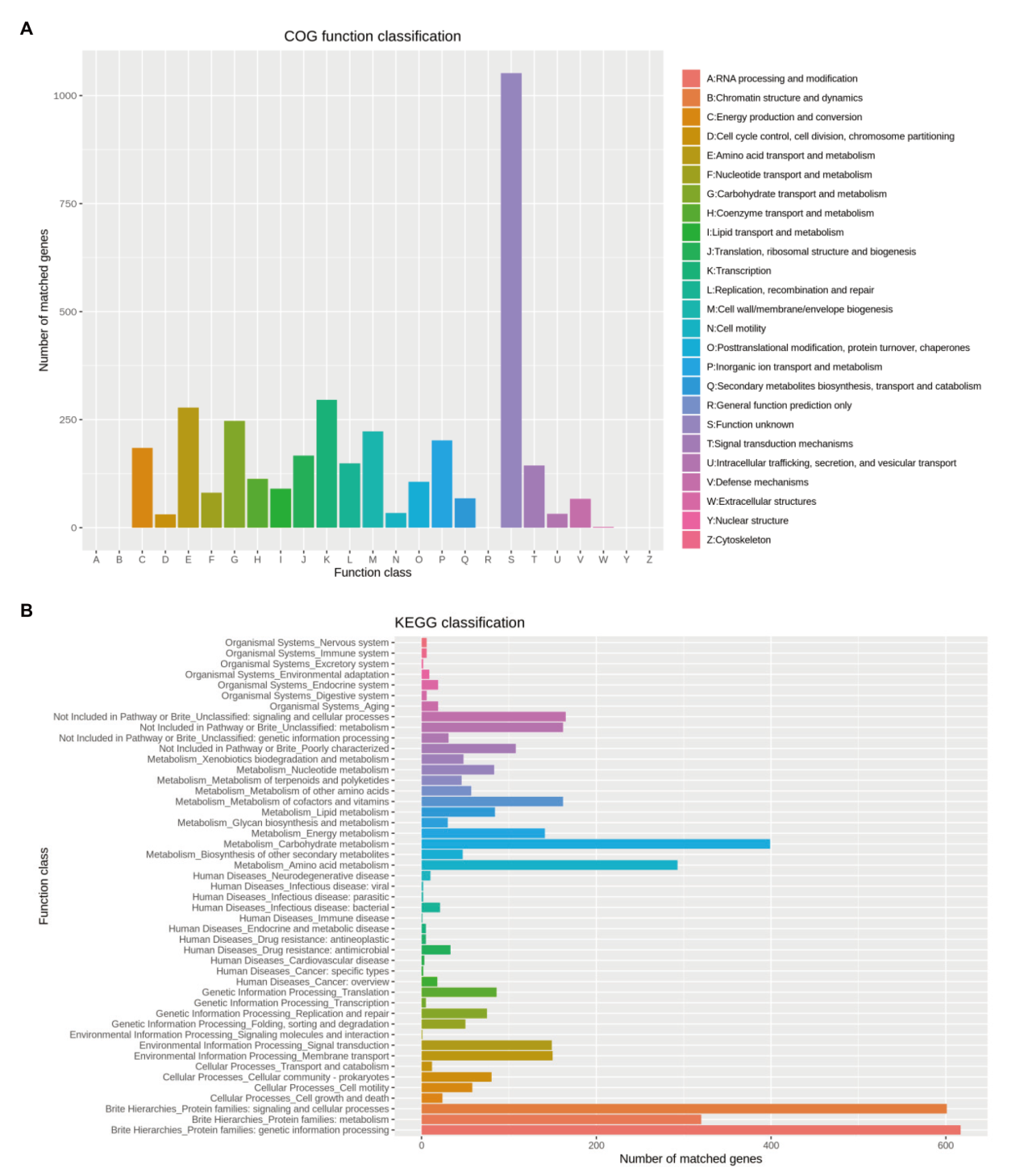
图7 菌株CNY01基因的COG和KEGG功能聚类分类 A:COG聚类分析;B:KEGG分类
Fig. 7 COG and KEGG functional classification analysis of genes in strain CNY01 A: COG functional classification; B: KEGG classification
| 位置Location | 长度Length/bp | 基因Gene | 产物Product |
|---|---|---|---|
| KCX77_05795 | 453 | nhaC | Na+/H+ antiporter NhaC |
| KCX77_09890 | 429 | / | Na+/H+ antiporter NhaC family protein |
| KCX77_12210 | 468 | nhaC | Na+/H+ antiporter NhaC |
| KCX77_15895 | 806 | / | Na+/H+ antiporter subunit A |
| KCX77_15910 | 493 | / | Na+/H+ antiporter subunit D |
| KCX77_15915 | 158 | / | Na+/H+ antiporter subunit E |
| KCX77_15925 | 124 | / | Na+/H+ antiporter subunit G |
| KCX77_16910 | 671 | / | Na+/H+ antiporter |
| KCX77_01700 | 418 | opuAA | Glycine/proline betaine ABC transporter |
| KCX77_01705 | 282 | opuAB | Glycine/proline betaine ABC transporter permease subunit OpuAB |
| KCX77_01805 | 303 | / | Proline dehydrogenase |
| KCX77_01815 | 504 | putP | Sodium/proline symporter PutP |
| KCX77_04225 | 492 | putP | Sodium/proline symporter PutP |
| KCX77_09365 | 564 | proS | Proline--tRNA ligase |
| KCX77_16595 | 302 | / | Proline dehydrogenase |
| KCX77_17080 | 381 | / | Betaine/proline/choline family ABC transporter ATP-binding protein |
| KCX77_00320 | 205 | / | General stress protein Ctc |
| KCX77_04635 | 115 | / | General stress protein |
| KCX77_15790 | 130 | yugI | General stress protein 13 |
| KCX77_19470 | 547 | katX | Catalase KatX |
| KCX77_02490 | 273 | / | Manganese catalase family protein |
| KCX77_05335 | 483 | katA | Catalase KatA |
| KCX77_12565 | 285 | / | Manganese catalase family protein |
| KCX77_08110 | 302 | / | Manganese catalase family protein |
| KCX77_19655 | 685 | / | Catalase |
| KCX77_07800 | 449 | / | TrkH family potassium uptake protein |
| KCX77_08320 | 221 | ktrC | Ktr system potassium transporter KtrC |
| KCX77_15595 | 222 | / | TrkA family potassium uptake protein |
| KCX77_15600 | 445 | / | TrkH family potassium uptake protein |
| KCX77_15755 | 328 | / | Potassium channel family protein |
| KCX77_19400 | 221 | / | TrkA family potassium uptake protein |
| KCX77_17200 | 394 | / | Phosphoglycerate kinase |
| KCX77_14540 | 585 | pyk | Pyruvate kinase |
表3 菌株CNY01的部分耐盐促生相关基因分析
Table 3 Analysis of some genes related to salt tolerance and growth promotion in strain CNY01
| 位置Location | 长度Length/bp | 基因Gene | 产物Product |
|---|---|---|---|
| KCX77_05795 | 453 | nhaC | Na+/H+ antiporter NhaC |
| KCX77_09890 | 429 | / | Na+/H+ antiporter NhaC family protein |
| KCX77_12210 | 468 | nhaC | Na+/H+ antiporter NhaC |
| KCX77_15895 | 806 | / | Na+/H+ antiporter subunit A |
| KCX77_15910 | 493 | / | Na+/H+ antiporter subunit D |
| KCX77_15915 | 158 | / | Na+/H+ antiporter subunit E |
| KCX77_15925 | 124 | / | Na+/H+ antiporter subunit G |
| KCX77_16910 | 671 | / | Na+/H+ antiporter |
| KCX77_01700 | 418 | opuAA | Glycine/proline betaine ABC transporter |
| KCX77_01705 | 282 | opuAB | Glycine/proline betaine ABC transporter permease subunit OpuAB |
| KCX77_01805 | 303 | / | Proline dehydrogenase |
| KCX77_01815 | 504 | putP | Sodium/proline symporter PutP |
| KCX77_04225 | 492 | putP | Sodium/proline symporter PutP |
| KCX77_09365 | 564 | proS | Proline--tRNA ligase |
| KCX77_16595 | 302 | / | Proline dehydrogenase |
| KCX77_17080 | 381 | / | Betaine/proline/choline family ABC transporter ATP-binding protein |
| KCX77_00320 | 205 | / | General stress protein Ctc |
| KCX77_04635 | 115 | / | General stress protein |
| KCX77_15790 | 130 | yugI | General stress protein 13 |
| KCX77_19470 | 547 | katX | Catalase KatX |
| KCX77_02490 | 273 | / | Manganese catalase family protein |
| KCX77_05335 | 483 | katA | Catalase KatA |
| KCX77_12565 | 285 | / | Manganese catalase family protein |
| KCX77_08110 | 302 | / | Manganese catalase family protein |
| KCX77_19655 | 685 | / | Catalase |
| KCX77_07800 | 449 | / | TrkH family potassium uptake protein |
| KCX77_08320 | 221 | ktrC | Ktr system potassium transporter KtrC |
| KCX77_15595 | 222 | / | TrkA family potassium uptake protein |
| KCX77_15600 | 445 | / | TrkH family potassium uptake protein |
| KCX77_15755 | 328 | / | Potassium channel family protein |
| KCX77_19400 | 221 | / | TrkA family potassium uptake protein |
| KCX77_17200 | 394 | / | Phosphoglycerate kinase |
| KCX77_14540 | 585 | pyk | Pyruvate kinase |
| [1] | 王佳丽, 黄贤金, 钟太洋, 等. 盐碱地可持续利用研究综述[J]. 地理学报, 2011, 66(5): 673-684. |
|
Wang JL, Huang XJ, Zhong TY, et al. Review on sustainable utilization of salt-affected land[J]. Acta Geogr Sin, 2011, 66(5): 673-684.
doi: 10.11821/xb201105010 |
|
| [2] | 魏博娴. 中国盐碱土的分布与成因分析[J]. 水土保持应用技术, 2012(6): 27-28. |
| Wei BX. Distribution and cause analysis of saline-alkali soil in China[J]. Technol Soil Water Conserv, 2012(6): 27-28. | |
| [3] | 赵娇. 基于菌群改善促进盐碱地耐受性植物生长的研究[D]. 济南: 山东大学, 2020. |
| Zhao J. Study on the improvement of flora to promote thegrowth of tolerant plants in saline-alkali land[D]. Jinan: Shandong University, 2020. | |
| [4] | 王文飞. 萎缩芽孢杆菌WU-9与辣椒互作缓解盐胁迫机制及菌株相关功能基因解析[D]. 石河子: 石河子大学, 2022. |
| Wang WF. Mechanism of relieving salt stress by interaction between Bacillus Atrophyta WU-9 and pepper and analysis of related functional genes of the strain[D]. Shihezi: Shihezi University, 2022. | |
| [5] | 郭英. 生防芽孢杆菌对作物耐盐性的影响及其抗盐机理的研究[D]. 济南: 山东师范大学, 2009. |
| Guo Y. Effects and mechanism of biocontrol strain of Bacillus subtilis on salt tolerance of plants[D]. Jinan: Shandong Normal University, 2009. | |
| [6] | 庞亚琴, 任彩婷, 徐秋曼. 解淀粉芽孢杆菌HM618对镉胁迫下小麦幼苗生长的影响[J]. 天津师范大学学报: 自然科学版, 2018, 38(4): 55-59. |
| Pang YQ, Ren CT, Xu QM. Effects of Bacillus amyloliquefaciens HM618 on the growth of wheat seedlings under cadmium stress[J]. J Tianjin Norm Univ Nat Sci Ed, 2018, 38(4): 55-59. | |
| [7] |
Sella SRBR, Vandenberghe LPS, Soccol CR. Bacillus atrophaeus: main characteristics and biotechnological applications - a review[J]. Crit Rev Biotechnol, 2015, 35(4): 533-545.
doi: 10.3109/07388551.2014.922915 pmid: 24963702 |
| [8] | Ma JJ, Wang CQ, Wang HD, et al. Analysis of the complete genome sequence of Bacillus atrophaeus GQJK17 reveals its biocontrol characteristics as a plant growth-promoting rhizobacterium[J]. Biomed Res Int, 2018, 2018: 9473542. |
| [9] |
Gordon RE, Smith NR. Aerobic sporeforming bacteria capable of growth at high temperatures[J]. J Bacteriol, 1949, 58(3): 327-341.
doi: 10.1128/jb.58.3.327-341.1949 pmid: 16561790 |
| [10] | Nakamura LK. Taxonomic relationship of black-pigmented Bacillus subtilis strains and a proposal for Bacillus atrophaeus sp. nov.[J]. International Journal of Systematic and Evolutionary Microbiology, 1989, 39(3):295-300. |
| [11] | Hou YL, Zeng WZ, Ao C, et al. Bacillus atrophaeus WZYH01 and Planococcus soli WZYH02 improve salt tolerance of maize(Zea mays L.) in saline soil[J]. Front Plant Sci, 2022, 13: 891372. |
| [12] | 韦廷舟, 文怡, 王超, 等. 一株产IAA芽孢杆菌ST37对油菜的耐盐促生作用[J]. 江苏农业科学, 2023, 51(6): 210-215. |
| Wei TZ, Wen Y, Wang C, et al. Effect of an IAA-producing Bacillus ST37 on salt tolerance and growth promotion of rape[J]. Jiangsu Agric Sci, 2023, 51(6): 210-215. | |
| [13] | 郇惠杰, 钟泓波, 雷芬芬, 等. 产蛋白酶海洋细菌的筛选、鉴定及发酵培养基的研究[J]. 食品工业科技, 2013, 34(24): 181-185. |
| Huan HJ, Zhong HB, Lei FF, et al. Study on isolation and identification of protease-producing marine bacteria and optimization of fermentation medium[J]. Sci Technol Food Ind, 2013, 34(24): 181-185. | |
| [14] | 张祥胜. 发酵液有效磷含量测定方法研究[J]. 湖州职业技术学院学报, 2008, 6(3): 1-3. |
| Zhang XS. A study of factors affecting the determined value by Mo-Sn-vc method of organic phosphobacteria[J]. J Huzhou Vocat Technol Coll, 2008, 6(3): 1-3. | |
| [15] | 李浩. 玉米、黄瓜根际促生菌组合优化及基因组测序[D]. 泰安: 山东农业大学, 2019. |
| Li H. Optimization of rhizosphere growth-promoting bacteria combination and genome sequencing of maize and cucumber[D]. Tai'an: Shandong Agricultural University, 2019. | |
| [16] | 马锦锦. 枸杞根际促生细菌筛选、培养基优化及基因组测序[D]. 泰安: 山东农业大学, 2018. |
| Ma JJ. Screening, Medium optimization and genome sequencing of PGPR from the rhizosphere of Lycium barbarum L.[D]. Tai'an: Shandong Agricultural University, 2018. | |
| [17] | 侯贞. 烟草根际促生细菌的筛选鉴定及发酵培养基优化[D]. 泰安: 山东农业大学, 2015. |
| Hou Z. Screening and identification of tobacco rhizosphere growth-promoting bacteria and optimization of fermentation medium[D]. Tai'an: Shandong Agricultural University, 2015. | |
| [18] | 杨亚男. 番茄根际促生菌的筛选及其培养基优化[D]. 泰安: 山东农业大学, 2017. |
| Yang YN. Screening of tomato rhizosphere growth-promoting bacteria and optimization of its culture medium[D]. Tai'an: Shandong Agricultural University, 2017. | |
| [19] |
Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging[J]. BMC Res Notes, 2016, 9: 88.
doi: 10.1186/s13104-016-1900-2 pmid: 26868221 |
| [20] | Luo RB, Liu BH, Xie YL, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler[J]. GigaScience, 2012, 1(1): 18. |
| [21] |
Chin CS, Peluso P, Sedlazeck FJ, et al. Phased diploid genome assembly with single-molecule real-time sequencing[J]. Nat Methods, 2016, 13(12): 1050-1054.
doi: 10.1038/NMETH.4035 |
| [22] | Koren S, Walenz BP, Berlin K, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation[J]. Genome Res, 2017, 27(5): 722-736. |
| [23] | Walker BJ, Abeel T, Shea T, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement[J]. PLoS One, 2014, 9(11): e112963. |
| [24] | 王欣. 植物根际促生细菌CNY01和BY2G20的耐盐功能及其对玉米的抗盐促生研究[D]. 泰安: 山东农业大学, 2021. |
| Wang X, The Salt Tolerance Function of Plant Growth-promoting Rhizobacteria CNY01 and BY2G20 and Their Anti-salt Promotion for Corn Growth[D]. Tai'an: Shandong Agricultural University, 2021. | |
| [25] | 索雲凯, 刘丽红, 张雷, 等. 解钾菌解钾作用研究进展[J]. 当代化工, 2021, 50(4): 924-929. |
| Suo YK, Liu LH, Zhang L, et al. Research progress of potassium solubilization by potassium solubilizing bacteria[J]. Contemp Chem Ind, 2021, 50(4): 924-929. | |
| [26] | Ayaz M, Ali Q, Farzand A, et al. Nematicidal volatiles from Bacillus atrophaeus GBSC56 promote growth and stimulate induced systemic resistance in tomato against Meloidogyne incognita[J]. Int J Mol Sci, 2021, 22(9): 5049. |
| [27] | 刘思靖. 萎缩芽孢杆菌次级代谢产物的研究进展[J]. 现代化工, 2019, 39(12): 48-51. |
| Liu SJ. Advances in secondary metabolites from Bacillus atrophaeus[J]. Mod Chem Ind, 2019, 39(12): 48-51. | |
| [28] |
戴宝, 扈进冬, 尹姗姗, 等. 响应面法优化萎缩芽孢杆菌BsR05发酵培养基[J]. 山东科学, 2017, 30(4): 31-37.
doi: 10.3976/j.issn.1002-4026.2017.04.006 |
| Dai B, Hu JD, Yin SS, et al. Optimization of fermentation medium composition for Bacillus atrophaeus BsR05 by response surface method[J]. Shandong Sci, 2017, 30(4): 31-37. | |
| [29] | 王敏. 萎缩芽孢杆菌XW2对苹果树腐烂病的防效评价[D]. 乌鲁木齐: 新疆农业大学, 2020. |
| Wang M. Evaluation on control effect of Bacillus atrophyta XW2 on apple tree rot[D]. Urumqi: Xinjiang Agricultural University, 2020. | |
| [30] |
胡玉婕, 朱秀玲, 丁延芹, 等. 芽孢杆菌的耐盐促生机制研究进展[J]. 生物技术通报, 2020, 36(9): 64-74.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0746 |
| Hu YJ, Zhu XL, Ding YQ, et al. Research progress on salt tolerance and growth-promoting mechanism of Bacillus[J]. Biotechnol Bull, 2020, 36(9): 64-74. | |
| [31] | Karim R, Bouchra B, Fatima G, et al. Plant NHX antiporters: from function to biotechnological application, with case study[J]. Curr Protein Pept Sci, 2021, 22(1): 60-73. |
| [32] | 赵东晓, 董亚茹, 孙景诗, 等. NaCl胁迫对胡麻种子萌发、幼苗生长及Na+/H+逆向转运蛋白基因表达的影响[J]. 山东农业科学, 2020, 52(7): 40-45. |
| Zhao DX, Dong YR, Sun JS, et al. Effects of NaCl stress on seed germination, seedling growth and Na+/H+ antiporter protein gene expression of Linum usitatissimum L[J]. Shandong Agric Sci, 2020, 52(7): 40-45. | |
| [33] |
张傲洁, 李青云, 宋文红, 等. 基于苯酚降解的粪产碱杆菌Alcaligenes faecalis JF101的全基因组分析[J]. 生物技术通报, 2023, 39(10): 292-303.
doi: 10.13560/j.cnki.biotech.bull.1985.2023-0281 |
| Zhang AJ, Li QY, Song WH, et al. Whole genome sequencing analysis of a phenol-degrading strain Alcaligenes faecalis JF101[J]. Biotechnol Bull, 2023, 39(10): 292-303. |
| [1] | 王佳玮, 李晨, 刘建利, 周世杰, 易嘉敏, 杨谨源, 康鹏. 内生真菌接种方式对青贮玉米幼苗生长的影响[J]. 生物技术通报, 2024, 40(4): 189-202. |
| [2] | 胡伊娃, 陈露. 玉米野生种基因组研究进展及应用[J]. 生物技术通报, 2024, 40(3): 14-24. |
| [3] | 常泸尹, 王中华, 李凤敏, 高梓源, 张辉红, 王祎, 李芳, 韩燕来, 姜瑛. 玉米根际多功能促生菌的筛选及其对冬小麦-夏玉米轮作体系产量提升效果[J]. 生物技术通报, 2024, 40(1): 231-242. |
| [4] | 王腾辉, 葛雯冬, 罗雅方, 范震宇, 王玉书. 基于极端混合池(BSA)全基因组重测序的羽衣甘蓝白色叶基因定位[J]. 生物技术通报, 2023, 39(9): 176-182. |
| [5] | 王宝宝, 王海洋. 理想株型塑造之于玉米耐密改良[J]. 生物技术通报, 2023, 39(8): 11-30. |
| [6] | 方澜, 黎妍妍, 江健伟, 成胜, 孙正祥, 周燚. 盘龙参内生真菌胞内细菌7-2H的分离鉴定和促生特性研究[J]. 生物技术通报, 2023, 39(8): 272-282. |
| [7] | 郭少华, 毛会丽, 刘征权, 付美媛, 赵平原, 马文博, 李旭东, 关建义. 一株鱼源致病性嗜水气单胞菌XDMG的全基因组测序及比较基因组分析[J]. 生物技术通报, 2023, 39(8): 291-306. |
| [8] | 张道磊, 甘雨军, 乐亮, 普莉. 玉米产量性状的表观遗传调控机制和育种应用[J]. 生物技术通报, 2023, 39(8): 31-42. |
| [9] | 冷燕, 马晓薇, 陈光, 任鹤, 李翔. 玉米高产竞赛助力中国玉米种业振兴[J]. 生物技术通报, 2023, 39(8): 4-10. |
| [10] | 王天依, 王荣焕, 王夏青, 张如养, 徐瑞斌, 焦炎炎, 孙轩, 王继东, 宋伟, 赵久然. 玉米矮秆基因与矮秆育种研究[J]. 生物技术通报, 2023, 39(8): 43-51. |
| [11] | 刘月娥, 徐田军, 蔡万涛, 吕天放, 张勇, 薛洪贺, 王荣焕, 赵久然. 我国玉米超高产研究现状与展望[J]. 生物技术通报, 2023, 39(8): 52-61. |
| [12] | 张勇, 徐田军, 吕天放, 邢锦丰, 刘宏伟, 蔡万涛, 刘月娥, 赵久然, 王荣焕. 种植密度对夏播玉米茎秆质量和根系表型性状的影响[J]. 生物技术通报, 2023, 39(8): 70-79. |
| [13] | 朱少喜, 金肇阳, 葛建镕, 王蕊, 王凤格, 路运才. 基于KASP平台的转基因玉米高通量特异性检测方法[J]. 生物技术通报, 2023, 39(6): 133-140. |
| [14] | 陈楠楠, 王春来, 蒋振忠, 焦鹏, 关淑艳, 马义勇. 玉米ZmDHN15基因在烟草中的遗传转化及抗冷性分析[J]. 生物技术通报, 2023, 39(4): 259-267. |
| [15] | 张志霞, 李天培, 曾虹, 朱稀贤, 杨天雄, 马斯楠, 黄磊. 冰冷杆菌PG-2的基因组测序及生物信息学分析[J]. 生物技术通报, 2023, 39(3): 290-300. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||