








生物技术通报 ›› 2024, Vol. 40 ›› Issue (7): 323-334.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0076
田彤彤1,2( ), 葛家振1, 高鹏程1, 李学瑞1, 宋国栋1, 郑福英1(
), 葛家振1, 高鹏程1, 李学瑞1, 宋国栋1, 郑福英1( ), 储岳峰1,2(
), 储岳峰1,2( )
)
收稿日期:2024-01-17
出版日期:2024-07-26
发布日期:2024-07-30
通讯作者:
郑福英,女,博士,研究员,研究方向:草食动物细菌病防控技术;E-mail: zhengfuying@caas.cn;作者简介:田彤彤,女,硕士,研究方向:预防兽医学;E-mail: t1123859519@163.com
基金资助:
TIAN Tong-tong1,2( ), GE Jia-zhen1, GAO Peng-cheng1, LI Xue-rui1, SONG Guo-dong1, ZHENG Fu-ying1(
), GE Jia-zhen1, GAO Peng-cheng1, LI Xue-rui1, SONG Guo-dong1, ZHENG Fu-ying1( ), CHU Yue-feng1,2(
), CHU Yue-feng1,2( )
)
Received:2024-01-17
Published:2024-07-26
Online:2024-07-30
摘要:
【目的】 全面了解绵羊肺炎支原体GH3-3株的基因序列,研究其潜在的致病机制及其复制、转录、翻译过程的调控机制。【方法】 采用体外培养,利用细菌基因组DNA提取试剂盒提取绵羊肺炎支原体GH3-3株基因组DNA,进行全基因组测序。【结果】 绵羊肺炎支原体GH3-3株基因组大小为1 060 772 bp,GC含量为29.66%,基因组组分分析后发现,GH3-3株的基因组含有730个编码基因,总长度为914 379 bp,平均长度为1 252.57 bp,占基因组全长的86.2%。串联重复序列共149个,总长为20 926 bp,占基因组全长的1.97%。微卫星DNA序列102个,tRNA 30个,rRNA 3个。在NR、SwissProt、GOG、KEGG、GO、CARD、CAZy、PHI、TCDB、RMS数据库中,分别有719、459、473、394、449、33、5、180、113、59个基因被注释;在VFDB数据库中,共注释到了76个毒力因子相关的基因。将基因组序列提交至NCBI网站,获得登录号为:PRJNA1051969。【结论】 获得了绵羊肺炎支原体GH3-3株完整的基因组信息,预测和注释了其基因的功能,明确了GH3-3株以及与国内外其他绵羊肺炎支原体菌株之间的遗传进化关系。
田彤彤, 葛家振, 高鹏程, 李学瑞, 宋国栋, 郑福英, 储岳峰. 绵羊肺炎支原体GH3-3株全基因组测序及生物信息学分析[J]. 生物技术通报, 2024, 40(7): 323-334.
TIAN Tong-tong, GE Jia-zhen, GAO Peng-cheng, LI Xue-rui, SONG Guo-dong, ZHENG Fu-ying, CHU Yue-feng. Whole Genome Sequencing and Bioinformatics Analysis of Mycoplasma ovipneumoniae GH3-3 Strain[J]. Biotechnology Bulletin, 2024, 40(7): 323-334.
| 菌株 Strain | 基因组大小 Genome size/bp | 编码基因数量 Gene number/个 | GC含量 GC content/% |
|---|---|---|---|
| GH 3-3株 | 1 060 772 | 730 | 29.66 |
| NXNK2203株 | 1 014 835 | 686 | 29.23 |
| NM2010株 | 1 084 159 | 888 | 29.14 |
| HHHT2017株 | 1 019 689 | 807 | 29.068 |
| SC01株 | 1 020 601 | 864 | 28.85 |
| WXB1株 | 1 084 159 | 888 | 29.14 |
| 150株 | 1 053 380 | 714 | 29.15 |
| 274株 | 1 081 404 | 747 | 28.82 |
| Y98株 | 1 021 486 | 819 | 29.21 |
表1 不同MO菌株基因组信息对比
Table 1 Comparison of genomic information of different MO strains
| 菌株 Strain | 基因组大小 Genome size/bp | 编码基因数量 Gene number/个 | GC含量 GC content/% |
|---|---|---|---|
| GH 3-3株 | 1 060 772 | 730 | 29.66 |
| NXNK2203株 | 1 014 835 | 686 | 29.23 |
| NM2010株 | 1 084 159 | 888 | 29.14 |
| HHHT2017株 | 1 019 689 | 807 | 29.068 |
| SC01株 | 1 020 601 | 864 | 28.85 |
| WXB1株 | 1 084 159 | 888 | 29.14 |
| 150株 | 1 053 380 | 714 | 29.15 |
| 274株 | 1 081 404 | 747 | 28.82 |
| Y98株 | 1 021 486 | 819 | 29.21 |

图6 Top10物种分布图 A: NR库Top10物种分布图;B: SwissProt库Top10物种分布图
Fig. 6 Distribution of top10 species A: Distribution of top10 species in NR database. B: Distribution of top10 species in SwissProt database
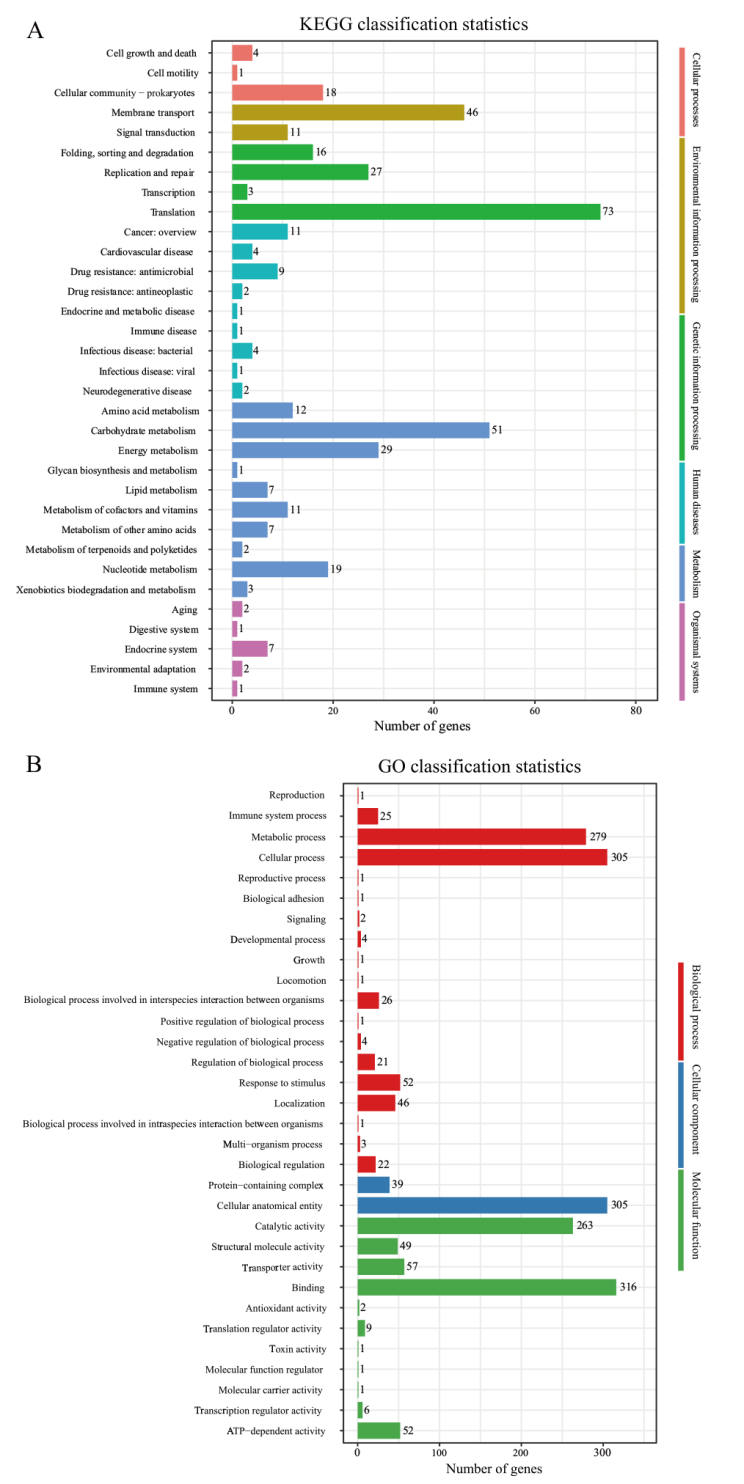
图8 KEGG、GO注释结果分类统计图 A:KEGG注释结果分类统计图;B:GO注释结果分类统计图
Fig. 8 Classification statistics diagrams of KEGG and GO annotation A: Classification statistics diagram of KEGG annotation. B: Classification statistics diagram of GO annotation
| 抗生素种类 Drug class | ARO登记号ARO accession | ARO名 ARO name |
|---|---|---|
| Aminoglycoside antibiotic | ARO:3005091 | RanA |
| Fluoroquinolone antibiotic | ARO:3003949 | efrB |
| Lincosamide antibiotic | ARO:3002882 | lmrD |
| ARO:3003746 | optrA | |
| ARO:3002825 | ErmY | |
| Macrolide antibiotic | ARO:3003949 | efrB |
| ARO:3003746 | optrA | |
| ARO:3002825 | ErmY | |
| ARO:3000535 | macB | |
| Mupirocin | ARO:3000510 | Staphylococcus aureus mupB conferring resistance to mupirocin |
| ARO:3000521 | Staphylococcus aureus mupA conferring resistance to mupirocin | |
| Nitroimidazole antibiotic | ARO:3003950 | msbA |
| Peptide antibiotic | ARO:3002987 | bcrA |
| ARO:3000501 | rpoB2 | |
| Tetracycline antibiotic | ARO:3004032 | tetA(46) |
| ARO:3000193 | tetT | |
| ARO:3002891 | otr(A) | |
| ARO:3003980 | tetA(58) | |
| ARO:3000556 | tet(44) | |
| ARO:3003980 | tetA(58) | |
| ARO:3003746 | optrA | |
| Rifamycin antibiotic | ARO:3003949 | efrB |
| ARO:3002825 | ErmY | |
| ARO:3000501 | rpoB2 | |
| Oxazolidinone antibiotic | ARO:3003746 | optrA |
| Pleuromutilin antibiotic | ARO:3003746 | optrA |
| Streptogramin antibiotic | ARO:3003746 | optrA |
| phenicol antibiotic | ARO:3003746 | optrA |
表2 耐药基因注释结果
Table 2 Annotation of drug resistance genes
| 抗生素种类 Drug class | ARO登记号ARO accession | ARO名 ARO name |
|---|---|---|
| Aminoglycoside antibiotic | ARO:3005091 | RanA |
| Fluoroquinolone antibiotic | ARO:3003949 | efrB |
| Lincosamide antibiotic | ARO:3002882 | lmrD |
| ARO:3003746 | optrA | |
| ARO:3002825 | ErmY | |
| Macrolide antibiotic | ARO:3003949 | efrB |
| ARO:3003746 | optrA | |
| ARO:3002825 | ErmY | |
| ARO:3000535 | macB | |
| Mupirocin | ARO:3000510 | Staphylococcus aureus mupB conferring resistance to mupirocin |
| ARO:3000521 | Staphylococcus aureus mupA conferring resistance to mupirocin | |
| Nitroimidazole antibiotic | ARO:3003950 | msbA |
| Peptide antibiotic | ARO:3002987 | bcrA |
| ARO:3000501 | rpoB2 | |
| Tetracycline antibiotic | ARO:3004032 | tetA(46) |
| ARO:3000193 | tetT | |
| ARO:3002891 | otr(A) | |
| ARO:3003980 | tetA(58) | |
| ARO:3000556 | tet(44) | |
| ARO:3003980 | tetA(58) | |
| ARO:3003746 | optrA | |
| Rifamycin antibiotic | ARO:3003949 | efrB |
| ARO:3002825 | ErmY | |
| ARO:3000501 | rpoB2 | |
| Oxazolidinone antibiotic | ARO:3003746 | optrA |
| Pleuromutilin antibiotic | ARO:3003746 | optrA |
| Streptogramin antibiotic | ARO:3003746 | optrA |
| phenicol antibiotic | ARO:3003746 | optrA |
| 毒力因子名称 VFDB name | 相关基因 Related genes |
|---|---|
| <alpha>-Hemolysin | hlyB |
| Acinetobactin | bauE |
| Alginate regulation | mucD |
| Capsule | cpsB、FTT 0793、rpe |
| Cereulide | cesC |
| ClpC | ClpC |
| Cya | cyaB |
| Cytolysin | cylB |
| Dot/Icm | lpg2936 |
| EF-Tu | tufA |
| ESX-1 | eccA1 |
| FbpABC | fbpC |
| FeoAB | feoB |
| Flagella | flhF |
| FupA | feoB |
| HitABC | hitC |
| HP-NAP | napA |
| IgA1 protease | zmpC |
| LOS | licA |
| LplA1 | lplA1 |
| MgtBC | mgtB |
| MsrAB | msrA/B(pilB) |
| P97/P102 paralog family | mhp107、mhp683、p102、p97、lppT、p216、mhp385 |
| T4SS effectors | CBU 1566 |
| TlyC | tlyC |
| TTSS | escN、yscN、pscN |
| Type 1 fimbriae | fimE |
表3 毒力因子注释结果
Table 3 Annotation of virulence factors
| 毒力因子名称 VFDB name | 相关基因 Related genes |
|---|---|
| <alpha>-Hemolysin | hlyB |
| Acinetobactin | bauE |
| Alginate regulation | mucD |
| Capsule | cpsB、FTT 0793、rpe |
| Cereulide | cesC |
| ClpC | ClpC |
| Cya | cyaB |
| Cytolysin | cylB |
| Dot/Icm | lpg2936 |
| EF-Tu | tufA |
| ESX-1 | eccA1 |
| FbpABC | fbpC |
| FeoAB | feoB |
| Flagella | flhF |
| FupA | feoB |
| HitABC | hitC |
| HP-NAP | napA |
| IgA1 protease | zmpC |
| LOS | licA |
| LplA1 | lplA1 |
| MgtBC | mgtB |
| MsrAB | msrA/B(pilB) |
| P97/P102 paralog family | mhp107、mhp683、p102、p97、lppT、p216、mhp385 |
| T4SS effectors | CBU 1566 |
| TlyC | tlyC |
| TTSS | escN、yscN、pscN |
| Type 1 fimbriae | fimE |
| [1] | Maksimović Z, Rifatbegović M, Loria GR, et al. Mycoplasma ovipneumoniae: a most variable pathogen[J]. Pathogens, 2022, 11(12): 1477. |
| [2] | Li Y, Jiang Z, Xue D, et al. Mycoplasma ovipneumoniae induces sheep airway epithelial cell apoptosis through an ERK signalling-mediated mitochondria pathway[J]. BMC microbiology. 2016, 16(1):222. |
| [3] |
Carmichael LE, St George TD, Sullivan ND, et al. Isolation, propagation, and characterization studies of an ovine Mycoplasma responsible for proliferative interstitial pneumonia[J]. Cornell Vet, 1972, 62(4): 654-679.
pmid: 4672899 |
| [4] | Balish M, Bertaccini A, Blanchard A, et al. Recommended rejection of the names Malacoplasma gen. nov., Mesomycoplasma gen. nov., Metamycoplasma gen. nov., Metamycoplasmataceae fam. nov., Mycoplasmoidaceae fam. nov., Mycoplasmoidales ord. nov., Mycoplasmoides gen. nov., Mycoplasmopsis gen. nov.[Gupta, Sawnani, Adeolu, Alnajar and Oren 2018]and all proposed species comb. nov. placed therein[J]. Int J Syst Evol Microbiol, 2019, 69(11): 3650-3653. |
| [5] | Gupta RS, Oren A. Necessity and rationale for the proposed name changes in the classification of Mollicutes species. Reply to: ‘Recommended rejection of the names Malacoplasma gen. nov., Mesomycoplasma gen. nov., Metamycoplasma gen. nov., Metamycoplasmataceae fam. nov., Mycoplasmoidaceae fam. nov., Mycoplasmoidales ord. nov., Mycoplasmoides gen. nov., Mycoplasmopsis gen. nov.[Gupta, Sawnani, Adeolu, Alnajar and Oren 2018]and all proposed species comb. nov. placed therein’, by M. Balish et al.(Int J Syst Evol Microbiol, 2019;69: 3650-3653)[J]. Int J Syst Evol Microbiol, 2020, 70(2): 1431-1438. |
| [6] | Gupta RS, Sawnani S, Adeolu M, et al. Phylogenetic framework for the phylum Tenericutes based on genome sequence data: proposal for the creation of a new order Mycoplasmoidales ord. nov., containing two new families Mycoplasmoidaceae fam. nov. and Metamycoplasmataceae fam. nov. harbouring Eperythrozoon, Ureaplasma and five novel Genera[J]. Antonie Van Leeuwenhoek, 2018, 111(9): 1583-1630. |
| [7] | 乌志勇. 绵羊肺炎支原体SY株分离鉴定及HSP70蛋白基因的克隆与分析[D]. 呼和浩特: 内蒙古农业大学, 2023. |
| Wu ZY. Isolation and identification of SY strain of Mycoplasma ovi-pneumoniae and gene cloning and analysis of HSP70 protein[D]. Hohhot: Inner Mongolia Agricultural University, 2023. | |
| [8] | 谭星. 绵羊肺炎支原体诱导巨噬细胞分泌的细胞外囊泡Lgals3bp蛋白促炎作用研究[D]. 银川: 宁夏大学, 2022. |
| Tan X. Proinflammatory effect of Lgals3bp protein in extracellular vesicles secreted by macrophages induced by Mycoplasma ovipneumoniae[D]. Yinchuan: Ningxia University, 2022. | |
| [9] | Gautier-Bouchardon AV. Antimicrobial resistance in Mycoplasma spp[J]. Microbiol Spectr, 2018, 6(4). |
| [10] | Ma CX, Li M, Peng H, et al. Mesomycoplasma ovipneumoniae from goats with respiratory infection: pathogenic characteristics, population structure, and genomic features[J]. BMC Microbiol, 2023, 23(1): 220. |
| [11] |
Kolmogorov M, Bickhart DM, Behsaz B, et al. metaFlye: scalable long-read metagenome assembly using repeat graphs[J]. Nat Methods, 2020, 17(11): 1103-1110.
doi: 10.1038/s41592-020-00971-x pmid: 33020656 |
| [12] | Walker BJ, Abeel T, Shea T, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement[J]. PLoS One, 2014, 9(11): e112963. |
| [13] | O’Leary NA, Wright MW, Brister JR, et al. Reference sequence(RefSeq)database at NCBI: current status, taxonomic expansion, and functional annotation[J]. Nucleic Acids Res, 2016, 44(D1): D733-D745. |
| [14] |
Boutet E, Lieberherr D, Tognolli M, et al. UniProtKB/swiss-prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view[J]. Methods Mol Biol, 2016, 1374: 23-54.
doi: 10.1007/978-1-4939-3167-5_2 pmid: 26519399 |
| [15] | Galperin MY, Wolf YI, Makarova KS, et al. COG database update: focus on microbial diversity, model organisms, and widespread pathogens[J]. Nucleic Acids Res, 2021, 49(D1): D274-D281. |
| [16] | Kanehisa M, Furumichi M, Sato Y, et al. KEGG for taxonomy-based analysis of pathways and genomes[J]. Nucleic Acids Res, 2023, 51(D1): D587-D592. |
| [17] | Consortium TGO. The Gene Ontology Resource: 20 years and still GOing strong[J]. Nucleic Acids Res, 2019, 47(D1): D330-D338. |
| [18] | Alcock BP, Huynh W, Chalil R, et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database[J]. Nucleic Acids Res, 2023, 51(D1): D690-D699. |
| [19] | Lombard V, Golaconda Ramulu H, Drula E, et al. The carbohydrate-active enzymes database(CAZy)in 2013[J]. Nucleic Acids Res, 2014, 42(Database issue): D490-D495. |
| [20] | Urban M, Cuzick A, Seager J, et al. PHI-base in 2022: a multi-species phenotype database for Pathogen-Host Interactions[J]. Nucleic Acids Res, 2022, 50(D1): D837-D847. |
| [21] | Chen LH, Zheng DD, Liu B, et al. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on[J]. Nucleic Acids Res, 2016, 44(D1): D694-D697. |
| [22] | Saier MH, Reddy VS, Moreno-Hagelsieb G, et al. The Transporter Classification Database(TCDB): 2021 update[J]. Nucleic Acids Res, 2021, 49(D1): D461-D467. |
| [23] | Roberts RJ, Vincze T, Posfai J, et al. REBASE: a database for DNA restriction and modification: enzymes, genes and genomes[J]. Nucleic Acids Res, 2023, 51(D1): D629-D630. |
| [24] |
Nielsen H. Predicting secretory proteins with SignalP[J]. Methods Mol Biol, 2017, 1611: 59-73.
doi: 10.1007/978-1-4939-7015-5_6 pmid: 28451972 |
| [25] |
Wang YJ, Huang H, Sun MA, et al. T3DB: an integrated database for bacterial type III secretion system[J]. BMC Bioinformatics, 2012, 13: 66.
doi: 10.1186/1471-2105-13-66 pmid: 22545727 |
| [26] |
Krogh A, Larsson B, von Heijne G, et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes[J]. J Mol Biol, 2001, 305(3): 567-580.
doi: 10.1006/jmbi.2000.4315 pmid: 11152613 |
| [27] |
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Mol Biol Evol, 2016, 33(7): 1870-1874.
doi: 10.1093/molbev/msw054 pmid: 27004904 |
| [28] | Xie JM, Chen YR, Cai GJ, et al. Tree Visualization By One Table(tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees[J]. Nucleic Acids Res, 2023, 51(W1): W587-W592. |
| [29] | Wang JD, Liu HY, Raheem A, et al. Exploring Mycoplasma ovipneumoniae NXNK2203 infection in sheep: insights from histopathology and whole genome sequencing[J]. BMC Vet Res, 2024, 20(1): 20. |
| [30] | Wang XH, Huang HB, Cheng C, et al. Complete genome sequence of Mycoplasma ovipneumoniae strain NM2010, which was isolated from a sheep in China[J]. J Integr Agric, 2014, 13(11): 2562-2563. |
| [31] | 韩瑞鑫. 绵羊肺炎支原体检测方法的建立及全基因组生物信息学分析[D]. 呼和浩特: 内蒙古农业大学, 2018. |
| Han RX. The establishment of detection method for Mycoplasma ovipneumoniae and analysis of it's genome bioinformatics[D]. Hohhot: Inner Mongolia Agricultural University, 2018. | |
| [32] | Yang FL, Tang C, Wang Y, et al. Genome sequence of Mycoplasma ovipneumoniae strain SC01[J]. J Bacteriol, 2011, 193(18): 5018. |
| [33] | 徐春光. 丝状支原体PG3株和绵羊肺炎支原体内蒙分离株基因组测序及可变表面脂蛋白基因克隆表达[D]. 呼和浩特: 内蒙古农业大学, 2013. |
| Xu CG. Genome sequencing of Mycoplasma mycoides Str.PG3and Mycoplasma ovipneumoniae from inner Mongolia and cloning and expression of variable surface lipoprotein genes[D]. Hohhot: Inner Mongolia Agricultural University, 2013. | |
| [34] | Hao HF, Maksimović Z, Ma LN, et al. Complete genome sequences of Mycoplasma ovipneumoniae strains 150 and 274, isolated from different regions in Bosnia and Herzegovina[J]. Microbiol Resour Announc, 2023, 12(3): e0001123. |
| [35] | Li M, Wang J, Hao XJ, et al. Complete Genome Sequence of Mycoplasma ovipneumoniae Strain Y98[C]. 哈尔滨: 第五届全国生物信息学与系统生物学学术大会论文集, 2012, 2:138-139. |
| Li M, Wang J, Hao XJ, et al. Complete Genome Sequence of Mycoplasma ovipneumoniae Strain Y98[C]. Harbin: Proceedings of the 5th National Conference on Bioinformatics and Systems Biology, 2012, 2:138-139. | |
| [36] |
Branton D, Deamer DW, Marziali A, et al. The potential and challenges of nanopore sequencing[J]. Nat Biotechnol, 2008, 26(10): 1146-1153.
doi: 10.1038/nbt.1495 pmid: 18846088 |
| [37] | 张金花. 绵羊肺炎支原体分离鉴定、病理组织学分析及靶器官主要细胞因子变化研究[D]. 银川: 宁夏大学, 2022. |
| Zhang JH. Isolation, identification and histopathological analysis of Mycoplasma ovipneumoniae and study on the change of main cytokines in target organs[D]. Yinchuan: Ningxia University, 2022. | |
| [38] | Liu W, Xiao SB, Li M, et al. Comparative genomic analyses of Mycoplasma hyopneumoniae pathogenic 168 strain and its high-passaged attenuated strain[J]. BMC Genomics, 2013, 14: 80. |
| [39] | 杨发龙, 汤承. 绵羊肺炎支原体毒力相关基因预测与分析[J]. 黑龙江畜牧兽医, 2013(19): 18-20. |
| Yang FL, Tang C. Prediction and analysis of the virulence- related genes in sheep Mycoplasma pneumoniae[J]. Heilongjiang Anim Sci Vet Med, 2013(19): 18-20. | |
| [40] | Mohamed AIS. 牛支原体HB0801株具有免疫原性分泌蛋白的鉴定[D]. 武汉: 华中农业大学, 2019. |
| Mohamed AIS. Identification of immunogenic secretory proteins of mycoplasma bovis HB0801[D]. Wuhan: Huazhong Agricultural University, 2019. |
| [1] | 周江鸿, 夏菲, 仲丽, 仇兰芬, 李广, 刘倩, 张国锋, 邵金丽, 李娜, 车少臣. 黄栌枯萎病拮抗细菌CCBC3-3-1的全基因组测序及比较基因组分析[J]. 生物技术通报, 2024, 40(7): 235-246. |
| [2] | 孙亚楠, 王春雪, 王欣, 杜秉海, 刘凯, 汪城墙. 萎缩芽孢杆菌CNY01的生防特性及其对玉米的抗盐促生作用[J]. 生物技术通报, 2024, 40(5): 248-260. |
| [3] | 王腾辉, 葛雯冬, 罗雅方, 范震宇, 王玉书. 基于极端混合池(BSA)全基因组重测序的羽衣甘蓝白色叶基因定位[J]. 生物技术通报, 2023, 39(9): 176-182. |
| [4] | 方澜, 黎妍妍, 江健伟, 成胜, 孙正祥, 周燚. 盘龙参内生真菌胞内细菌7-2H的分离鉴定和促生特性研究[J]. 生物技术通报, 2023, 39(8): 272-282. |
| [5] | 郭少华, 毛会丽, 刘征权, 付美媛, 赵平原, 马文博, 李旭东, 关建义. 一株鱼源致病性嗜水气单胞菌XDMG的全基因组测序及比较基因组分析[J]. 生物技术通报, 2023, 39(8): 291-306. |
| [6] | 张志霞, 李天培, 曾虹, 朱稀贤, 杨天雄, 马斯楠, 黄磊. 冰冷杆菌PG-2的基因组测序及生物信息学分析[J]. 生物技术通报, 2023, 39(3): 290-300. |
| [7] | 和梦颖, 刘文彬, 林震鸣, 黎尔彤, 汪洁, 金小宝. 一株抗革兰阳性菌的戈登氏菌WA4-43全基因组测序与分析[J]. 生物技术通报, 2023, 39(2): 232-242. |
| [8] | 吴莉丹, 冉雪琴, 牛熙, 黄世会, 李升, 王嘉福. 猪源致病性大肠杆菌基因组比较与毒力因子分析[J]. 生物技术通报, 2023, 39(12): 287-299. |
| [9] | 张傲洁, 李青云, 宋文红, 颜少慧, 唐爱星, 刘幽燕. 基于苯酚降解的粪产碱杆菌Alcaligenes faecalis JF101的全基因组分析[J]. 生物技术通报, 2023, 39(10): 292-303. |
| [10] | 王帅, 吕鸿睿, 张昊, 吴占文, 肖翠红, 孙冬梅. 解磷菌PSB-R全基因组测序鉴定及其解磷特性分析[J]. 生物技术通报, 2023, 39(1): 274-283. |
| [11] | 鲁兆祥, 王夕冉, 连新磊, 廖晓萍, 刘雅红, 孙坚. 基于功能宏基因组学挖掘抗生素耐药基因研究进展[J]. 生物技术通报, 2022, 38(9): 17-27. |
| [12] | 刘理慧, 储锦华, 隋雨欣, 陈杨, 程古月. 沙门氏菌中主要毒力因子的研究进展[J]. 生物技术通报, 2022, 38(9): 72-83. |
| [13] | 张泽颖, 范清锋, 邓云峰, 韦廷舟, 周正富, 周建, 王劲, 江世杰. 一株高产脂肪酶菌株WCO-9全基因组测序及比较基因组分析[J]. 生物技术通报, 2022, 38(10): 216-225. |
| [14] | 薛清, 杜虹锐, 薛会英, 王译浩, 王暄, 李红梅. 苜蓿滑刃线虫线粒体基因组及其系统发育研究[J]. 生物技术通报, 2021, 37(7): 98-106. |
| [15] | 陈体强, 徐晓兰, 石林春, 钟礼义. 紫芝栽培品种‘武芝2号’(‘紫芝S2’)全基因组测序及分析[J]. 生物技术通报, 2021, 37(11): 42-56. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||