








生物技术通报 ›› 2024, Vol. 40 ›› Issue (8): 152-163.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0252
刘丹丹( ), 王雷刚, 孙明慧, 焦小雨, 吴琼, 王文杰(
), 王雷刚, 孙明慧, 焦小雨, 吴琼, 王文杰( )
)
收稿日期:2024-03-15
出版日期:2024-08-26
发布日期:2024-07-30
通讯作者:
王文杰,男,研究员,研究方向:茶树品种、茶叶加工与品质;E-mail: wwj00@126.com作者简介:刘丹丹,女,硕士,助理研究员,研究方向:茶树种质资源与品种选育;E-mail: 1653082943@qq.com
基金资助:
LIU Dan-dan( ), WANG Lei-gang, SUN Ming-hui, JIAO Xiao-yu, WU Qiong, WANG Wen-jie(
), WANG Lei-gang, SUN Ming-hui, JIAO Xiao-yu, WU Qiong, WANG Wen-jie( )
)
Received:2024-03-15
Published:2024-08-26
Online:2024-07-30
摘要:
【目的】海藻糖-6-磷酸合成酶(trehalose-6-phosphate synthase, TPS)是植物体内海藻糖生物合成通路中的关键酶。对茶树[Camellia sinensis(L.)O. Kuntze]TPS基因家族进行鉴定和分析,并研究其在逆境胁迫下的表达模式,为开展TPS基因功能验证提供基础。【方法】利用生物信息学方法鉴定茶树CsTPS基因家族成员,分析其编码的蛋白理化特性、保守基序与结构域、染色体定位、系统进化关系和启动子顺式作用元件等生物学信息,利用转录组数据结合RT-qPCR技术分析CsTPS家族成员在干旱、盐和低温胁迫下的表达模式。【结果】在茶树基因组中鉴定到16个TPS基因家族成员,系统发育分析将它们划分为Class I和Class II两个亚族。同一亚族成员具有相似的motif组成和基因结构。共线性与进化分析表明,CsTPS基因在茶树基因组内发生了基因复制事件,且它们在进化过程中经历了纯化选择作用。RT-qPCR结果显示,6个候选基因CsTPS3、CsTPS5、CsTPS6、CsTPS9、CsTPS11和CsTPS14均被PEG、NaCl和低温胁迫诱导,且表达模式与转录组结果一致。【结论】从茶树全基因组中系统鉴定出16个TPS家族成员。茶树CsTPS3、CsTPS5、CsTPS6、CsTPS9、CsTPS11和CsTPS14基因对干旱、盐和低温胁迫均有响应,但其敏感性水平各不相同。
刘丹丹, 王雷刚, 孙明慧, 焦小雨, 吴琼, 王文杰. 茶树海藻糖-6-磷酸合成酶(TPS)基因家族鉴定与表达分析[J]. 生物技术通报, 2024, 40(8): 152-163.
LIU Dan-dan, WANG Lei-gang, SUN Ming-hui, JIAO Xiao-yu, WU Qiong, WANG Wen-jie. Genome-wide Identification and Expression Pattern Profiling of the Trehalose-6-phosphate Synthase(TPS)Gene Family in Tea Plant(Camellia sinensis)[J]. Biotechnology Bulletin, 2024, 40(8): 152-163.
| 基因名称Gene name | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') |
|---|---|---|
| β-actin | GCCATCTTTGATTGGAATGG | GGTGCCACAACCTTGATCTT |
| CsTPS3 | TTCTCACAGAGAGTGATTGAGGTG | GGACTATGGAGAAAGAACCCCATT |
| CsTPS5 | TTCTCACAGAGAGTGATTGAGGTG | GGACTATGGAGAAAGAACCCCATT |
| CsTPS6 | TGTCTTTATTGTTAGTGGGAGGGG | CACAATTTCCTTCCACTCAAGGTC |
| CsTPS9 | GCAAGATATGGAGAGAACTTGTGC | TGGGGCATTACAGTACCATCATAG |
| CsTPS11 | GGTTACCTGTCTCTGCAGTTAGAA | CAAAGCTTTAGTAAGTGCCCTCTG |
| CsTPS14 | GCCCTGATGATGATTTTGTATGGG | TTCAACTCTGACAGGTAGTGTACG |
表1 Real-time PCR 引物信息
Table 1 Primer sequences for real-time PCR
| 基因名称Gene name | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') |
|---|---|---|
| β-actin | GCCATCTTTGATTGGAATGG | GGTGCCACAACCTTGATCTT |
| CsTPS3 | TTCTCACAGAGAGTGATTGAGGTG | GGACTATGGAGAAAGAACCCCATT |
| CsTPS5 | TTCTCACAGAGAGTGATTGAGGTG | GGACTATGGAGAAAGAACCCCATT |
| CsTPS6 | TGTCTTTATTGTTAGTGGGAGGGG | CACAATTTCCTTCCACTCAAGGTC |
| CsTPS9 | GCAAGATATGGAGAGAACTTGTGC | TGGGGCATTACAGTACCATCATAG |
| CsTPS11 | GGTTACCTGTCTCTGCAGTTAGAA | CAAAGCTTTAGTAAGTGCCCTCTG |
| CsTPS14 | GCCCTGATGATGATTTTGTATGGG | TTCAACTCTGACAGGTAGTGTACG |
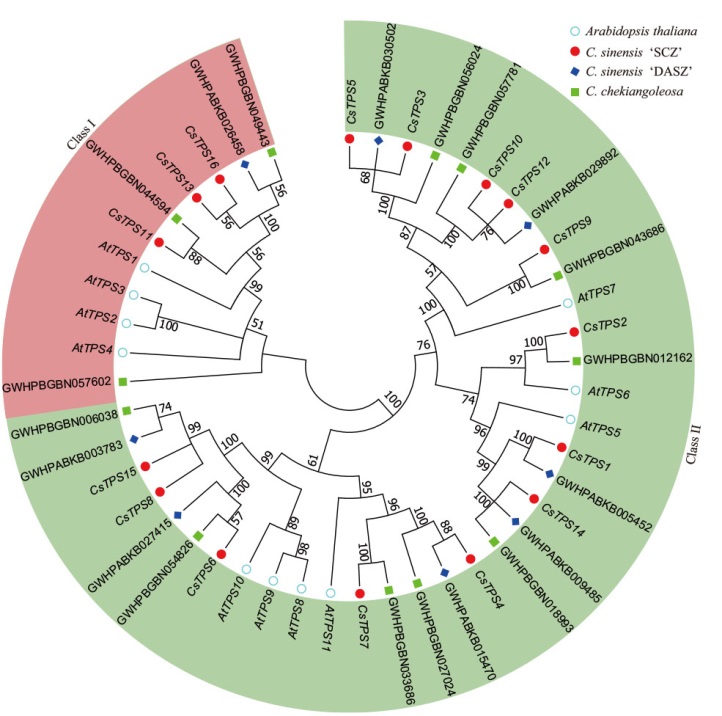
图1 拟南芥、舒茶早、云南野茶和浙江红山茶的TPS基因家族系统进化关系
Fig. 1 Phylogenetic relationship of TPS genes in A. thali-ana, C. sinensis ‘SCZ’, C. sinensis ‘DASZ’ and C. chekiangoleosa
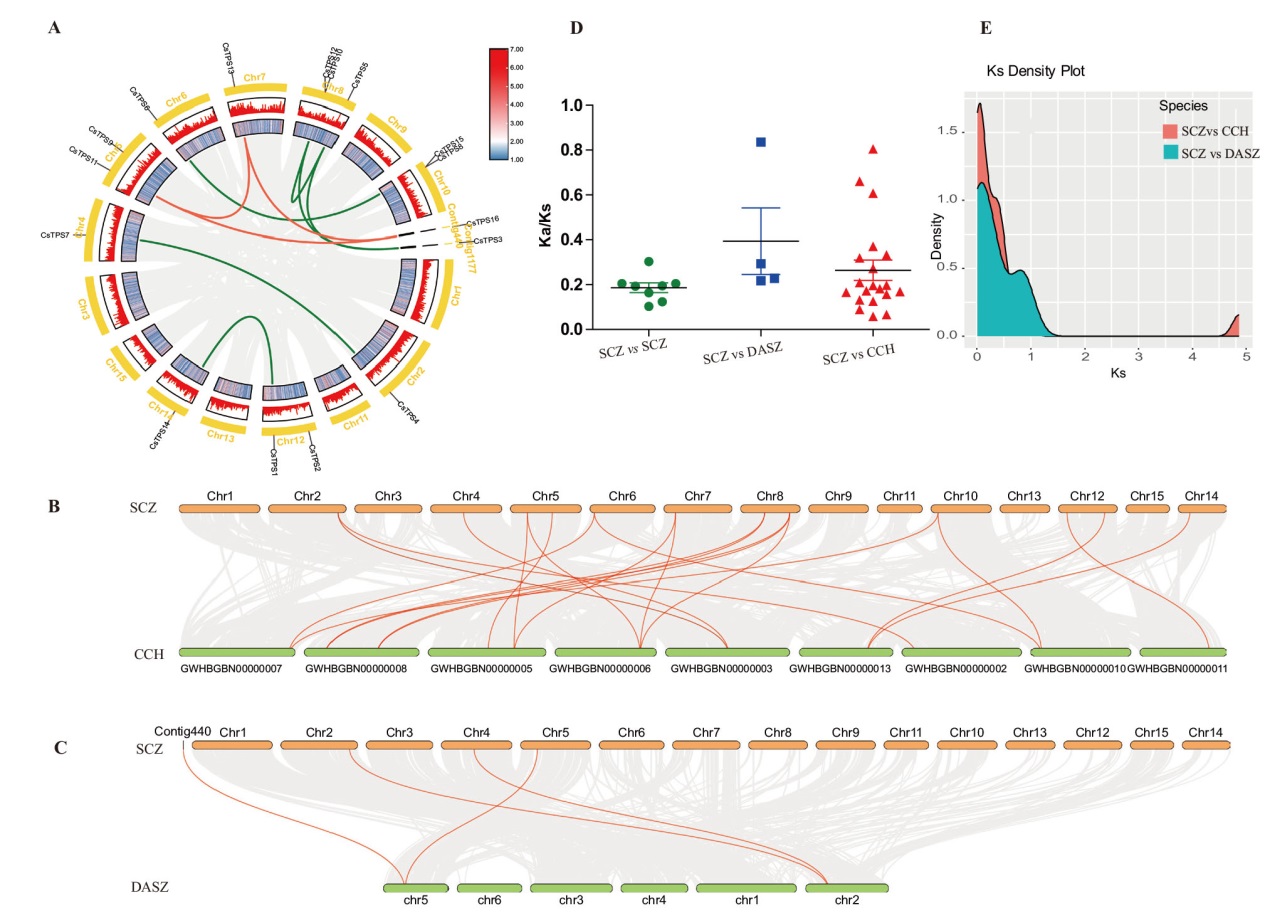
图5 山茶属植物TPS基因共线性与进化选择压力分析 A:茶树TPS基因共线性分析,基因组内共线性用灰色线表示,CsTPS基因家族Class I与Class II亚族成员之间的共线性分别用橙色线和绿色线表示;B:舒茶早(SCZ)与浙江红山茶(CCH)之间TPS基因共线性分析;C:舒茶早与云南野茶(DASZ)之间TPS基因共线性分析;D:Ka/Ks分析;E:3种山茶属植物同源基因对Ks的密度分布
Fig. 5 Analysis of syntenic and Ka/Ks of TPS genes in Camellia plants A: Syntenic analysis of TPS genes in C. sinensis. The inner gray lines indicate all the collinearity relationships in the whole genome, and the inner orange and green lines indicate the collinearity relationships of Class I and Class II subfamilies, respectively. B: Syntenic analysis of TPS genes between C. sinensis ‘SCZ’ and C. chekiangoleosa. C: Syntenic analysis of TPS genes between C. sinensis ‘SCZ’ and C. sinensis ‘DASZ’. D: Analysis of Ka/Ks. E: Density distribution of Ks for paralogous gene pairs of the three species of Camellia plants
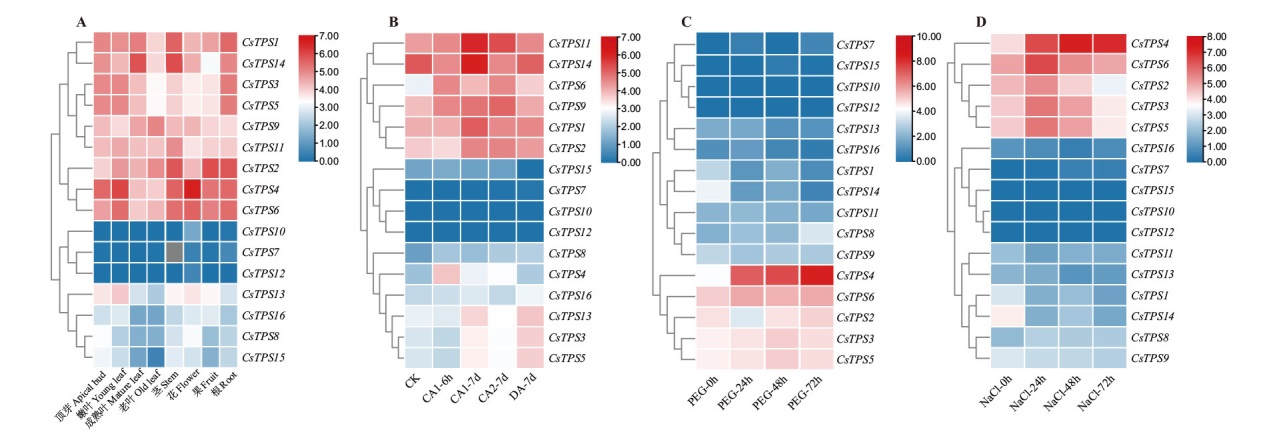
图6 茶树TPS基因转录组表达谱 A、C和D图分别表示CsTPS基因在不同组织、25% PEG胁迫和200 mmol/L NaCl胁迫下的表达模式;B:CsTPS基因在低温驯化下的表达模式(CK:白天25℃,晚上20℃,12 h光周期,70%湿度条件下生长1个月;CA1-6h:10℃处理6 h;CA1-7d:白天10℃,晚上4℃处理7 d;CA2-7d:白天4℃,晚上0℃处理7 d;DA-7d:白天25℃,晚上20℃处理7 d);热图体现的log2(FPKM+1)值
Fig. 6 Transcriptome expression profiling of TPS genes in C. sinensis A, C and D graphs indicate the expression patterns of CsTPS genes in different tissues, 25% PEG stress and 200 mmol/L NaCl stress, respectively. B: Expression pattern of CsTPS gene under low temperature domestication(CK: 1 month of growth at 25℃ during the day, 20℃ at night, 12 h photoperiod and 70% humidity; CA1-6h: 6 h treatment at 10℃; CA1-7d: 10℃ during the day, 4℃ at night treatment for 7 d; CA2-7d: 4℃ during the day and 0℃ at night for 7 d; DA-7d: 25℃ during the day and 20℃ at night for 7 d). The log2(FPKM+1)value reflects in the heatmap
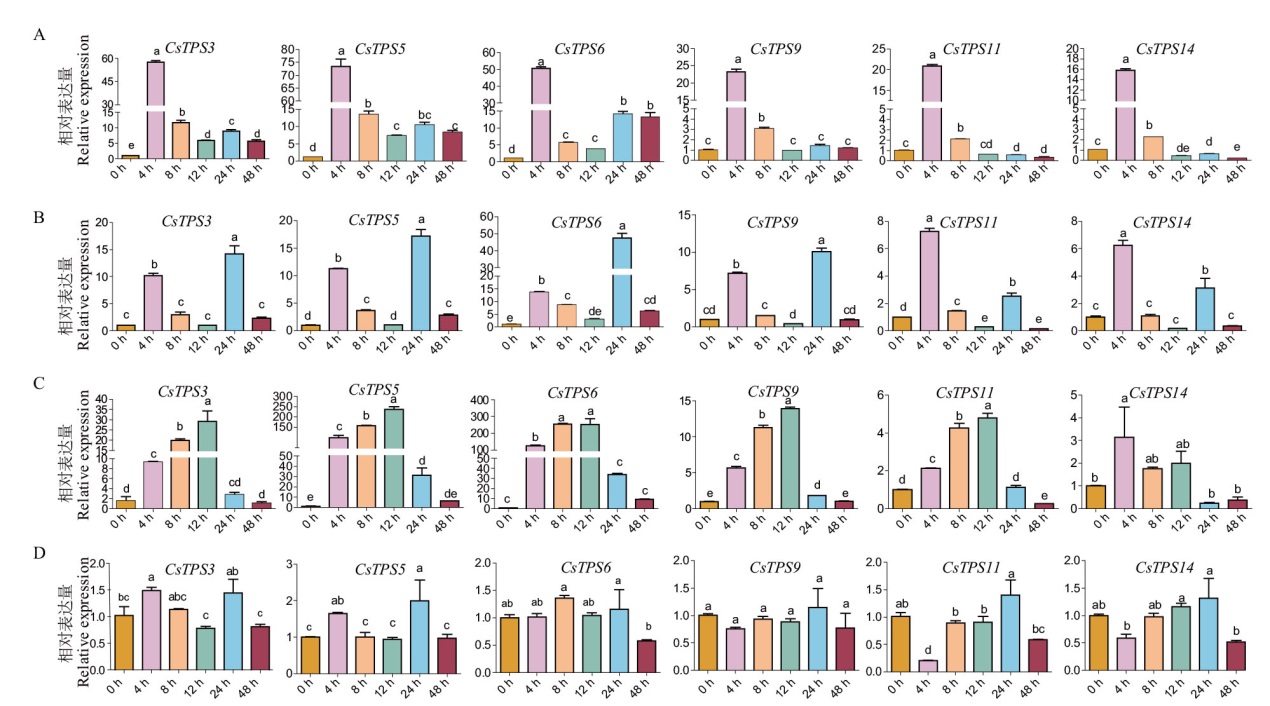
图7 不同胁迫下CsTPSs基因的表达分析 A:20% PEG处理下CsTPSs基因的表达谱;B:200 mmol/L NaCl处理下CsTPSs基因的表达谱;C:4℃处理下CsTPSs基因的表达谱;D:24℃下CsTPSs基因生物钟表达谱;误差线代表3次独立生物学重复标准偏差,不同小写字母代表同一种处理不同时间点差异显著(P<0.05)
Fig. 7 Relative expressions of CsTPS genes under various stress A: Gene expression profiles of CsTPSs under 20% PEG. B: Gene expression profiles of CsTPSs under 200 mmol/L NaCl. C: Gene expression profiles of CsTPSs under 4℃ treatment. D: Circadian expression profiling of the CsTPS genes at 24℃. Error bars indicate the standard deviation of three independent biological replicates, different lowercase letters in indicate significant differences at different time of the same treatment(P<0.05)
| [1] | Liu Y, Lee J, Mansfield KM, et al. Trehalose glycopolymer enhances both solution stability and pharmacokinetics of a therapeutic protein[J]. Bioconjug Chem, 2017, 28(3): 836-845. |
| [2] | Acosta-Pérez P, Camacho-Zamora BD, Espinoza-Sánchez EA, et al. Characterization of trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase genes and analysis of its differential expression in maize(Zea mays)seedlings under drought stress[J]. Plants, 2020, 9(3): 315. |
| [3] |
Márquez-Escalante JA, Figueroa-Soto CG, Valenzuela-Soto EM. Isolation and partial characterization of trehalose 6-phosphate synthase aggregates from Selaginella lepidophylla plants[J]. Biochimie, 2006, 88(10): 1505-1510.
pmid: 16828951 |
| [4] | Fichtner F, Lunn JE. The role of trehalose 6-phosphate(Tre6P)in plant metabolism and development[J]. Annu Rev Plant Biol, 2021, 72: 737-760. |
| [5] |
Vogel G, Aeschbacher RA, Müller J, et al. Trehalose-6-phosphate phosphatases from Arabidopsis thaliana: identification by functional complementation of the yeast tps2 mutant[J]. Plant J, 1998, 13(5): 673-683.
doi: 10.1046/j.1365-313x.1998.00064.x pmid: 9681009 |
| [6] |
Blázquez MA, Santos E, Flores CL, et al. Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase[J]. Plant J, 1998, 13(5): 685-689.
doi: 10.1046/j.1365-313x.1998.00063.x pmid: 9681010 |
| [7] |
Vandesteene L, López-Galvis L, Vanneste K, et al. Expansive evolution of the trehalose-6-phosphate phosphatase gene family in Arabidopsis[J]. Plant Physiol, 2012, 160(2): 884-896.
doi: 10.1104/pp.112.201400 pmid: 22855938 |
| [8] |
Ponnu J, Wahl V, Schmid M. Trehalose-6-phosphate: connecting plant metabolism and development[J]. Front Plant Sci, 2011, 2: 70.
doi: 10.3389/fpls.2011.00070 pmid: 22639606 |
| [9] | Figueroa CM, Feil R, Ishihara H, et al. Trehalose 6-phosphate coordinates organic and amino acid metabolism with carbon availability[J]. Plant J, 2016, 85(3): 410-423. |
| [10] | Lunn JE, Delorge I, Figueroa CM, et al. Trehalose metabolism in plants[J]. Plant J, 2014, 79(4): 544-567. |
| [11] | Avonce N, Wuyts J, Verschooten K, et al. The Cytophagahutchin-soniiChTPSP: first characterized bifunctional TPS-TPP protein as putative ancestor of all eukaryotic trehalose biosynthesis proteins[J]. Mol Biol Evol, 2010, 27(2): 359-369. |
| [12] |
Paul MJ, Primavesi LF, Jhurreea D, et al. Trehalose metabolism and signaling[J]. Annu Rev Plant Biol, 2008, 59: 417-441.
doi: 10.1146/annurev.arplant.59.032607.092945 pmid: 18257709 |
| [13] |
Miranda JA, Avonce N, Suárez R, et al. A bifunctional TPS-TPP enzyme from yeast confers tolerance to multiple and extreme abiotic-stress conditions in transgenic Arabidopsis[J]. Planta, 2007, 226(6): 1411-1421.
pmid: 17628825 |
| [14] | Chary SN, Hicks GR, Choi YG, et al. Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis[J]. Plant Physiol, 2008, 146(1): 97-107. |
| [15] | Satoh-Nagasawa N, Nagasawa N, Malcomber S, et al. A trehalose metabolic enzyme controls inflorescence architecture in maize[J]. Nature, 2006, 441(7090): 227-230. |
| [16] |
Eastmond PJ, Spielman M, et al. Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation[J]. Plant J, 2002, 29(2): 225-235.
doi: 10.1046/j.1365-313x.2002.01220.x pmid: 11851922 |
| [17] | Gómez LD, Gilday A, Feil R, et al. AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells[J]. Plant J, 2010, 64(1): 1-13. |
| [18] | Schluepmann H, Smeekens SCM. Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering[J]. Plant Physiol, 2004, 135(2): 969-977. |
| [19] |
Vandesteene L, Ramon M, Le Roy K, et al. A single active trehalose-6-P synthase(TPS)and a family of putative regulatory TPS-like proteins in Arabidopsis[J]. Mol Plant, 2010, 3(2): 406-419.
doi: 10.1093/mp/ssp114 pmid: 20100798 |
| [20] | Zang BS, Li HW, Li WJ, et al. Analysis of trehalose-6-phosphate synthase(TPS)gene family suggests the formation of TPS complexes in rice[J]. Plant Mol Biol, 2011, 76(6): 507-522. |
| [21] | Gao YH, Yang XY, Yang X, et al. Characterization and expression pattern of the trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase gene families in Populus[J]. Int J Biol Macromol, 2021, 187: 9-23. |
| [22] | Xu YC, Wang YJ, Mattson N, et al. Genome-wide analysis of the Solanum tuberosum(potato)trehalose-6-phosphate synthase(TPS)gene family: evolution and differential expression during development and stress[J]. BMC Genomics, 2017, 18(1): 926. |
| [23] | Du LS, Qi SY, Ma JJ, et al. Identification of TPS family members in apple(Malus×domestica Borkh.)and the effect of sucrose sprays on TPS expression and floral induction[J]. Plant PhysiolBiochem, 2017, 120: 10-23. |
| [24] | Dan YY, Niu Y, Wang CL, et al. Genome-wide identification and expression analysis of the trehalose-6-phosphate synthase(TPS)gene family in cucumber(Cucumis sativus L.)[J]. PeerJ, 2021, 9: e11398. |
| [25] |
Delorge I, Janiak M, Carpentier S, et al. Fine tuning of trehalose biosynthesis and hydrolysis as novel tools for the generation of abiotic stress tolerant plants[J]. Front Plant Sci, 2014, 5: 147.
doi: 10.3389/fpls.2014.00147 pmid: 24782885 |
| [26] | Ramon M, De Smet I, Vandesteene L, et al. Extensive expression regulation and lack of heterologous enzymatic activity of the Class II trehalose metabolism proteins from Arabidopsis thaliana[J]. Plant Cell Environ, 2009, 32(8): 1015-1032. |
| [27] | Avonce N, Leyman B, Mascorro-Gallardo JO, et al. The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling[J]. Plant Physiol, 2004, 136(3): 3649-3659. |
| [28] | Almeida AM, Villalobos E, Araújo SS, et al. Transformation of tobacco with an Arabidopsis thaliana gene involved in trehalose biosynthesis increases tolerance to several abiotic stresses[J]. Euphytica, 2005, 146(1): 165-176. |
| [29] | Li HW, Zang BS, Deng XW, et al. Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice[J]. Planta, 2011, 234(5): 1007-1018. |
| [30] | Tian LF, Xie ZJ, Lu CQ, et al. The trehalose-6-phosphate synthase TPS5 negatively regulates ABA signaling in Arabidopsis thaliana[J]. Plant Cell Rep, 2019, 38(8): 869-882. |
| [31] | 丁菲, 庞磊, 李叶云, 等. 茶树海藻糖-6-磷酸合成酶基因(CsTPS)的克隆及表达分析[J]. 农业生物技术学报, 2012, 20(11): 1253-1261. |
| Ding F, Pang L, Li YY, et al. Cloning and expression analysis of trehalose-6-phosphate synthase gene(CsTPS)from tea plant(Camellia sinensis(L.) O. kuntz)[J]. J Agric Biotechnol, 2012, 20(11): 1253-1261. | |
| [32] | Chen CJ, Wu Y, Li JW, et al. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining[J]. Mol Plant, 2023, 16(11): 1733-1742. |
| [33] | Wei CL, Yang H, Wang SB, et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality[J]. Proc Natl Acad Sci USA, 2018, 115(18): E4151-E4158. |
| [34] | Zhang Q, Cai MC, Yu XM, et al. Transcriptome dynamics of Camellia sinensis in response to continuous salinity and drought stress[J]. Tree Genet Genomes, 2017, 13(4): 78. |
| [35] | Li YY, Wang XW, Ban QY, et al. Comparative transcriptomic analysis reveals gene expression associated with cold adaptation in the tea plant Camellia sinensis[J]. BMC Genomics, 2019, 20(1): 624. |
| [36] | Cao QH, Lv WY, Jiang H, et al. Genome-wide identification of glutathione S-transferase gene family members in tea plant(Camellia sinensis)and their response to environmental stress[J]. Int J Biol Macromol, 2022, 205: 749-760. |
| [37] | Li PH, Xia EH, Fu JM, et al. Diverse roles of MYB transcription factors in regulating secondary metabolite biosynthesis, shoot development, and stress responses in tea plants(Camellia sinensis)[J]. Plant J, 2022, 110(4): 1144-1165. |
| [38] |
Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution[J]. Trends Genet, 2002, 18(9): 486.
doi: 10.1016/s0168-9525(02)02722-1 pmid: 12175810 |
| [39] | Song J, Gao ZH, Huo XM, et al. Genome-wide identification of the auxin response factor(ARF)gene family and expression analysis of its role associated with pistil development in Japanese apricot(Prunus mume Sieb. et Zucc)[J]. Acta Physiol Plant, 2015, 37(8): 145. |
| [40] | Xia EH, Zhang HB, Sheng J, et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis[J]. Mol Plant, 2017, 10(6): 866-877. |
| [41] | Shen TF, Huang B, Xu M, et al. The reference genome of Camellia chekiangoleosa provides insights into camellia evolution and tea oil biosynthesis[J]. Hortic Res, 2022, 9: uhab083. |
| [42] | Doi K, Hosaka A, Nagata T, et al. Development of a novel data mining tool to find cis-elements in rice gene promoter regions[J]. BMC Plant Biol, 2008, 8(1): 20. |
| [43] |
Gómez-Porras JL, Riaño-Pachón DM, Dreyer I, et al. Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice[J]. BMC Genomics, 2007, 8: 260.
pmid: 17672917 |
| [44] |
Karim S, Aronsson H, Ericson H, et al. Improved drought tolerance without undesired side effects in transgenic plants producing trehalose[J]. Plant Mol Biol, 2007, 64(4): 371-386.
doi: 10.1007/s11103-007-9159-6 pmid: 17453154 |
| [45] |
Garg AK, Kim JK, Owens TG, et al. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses[J]. Proc Natl Acad Sci USA, 2002, 99(25): 15898-15903.
pmid: 12456878 |
| [46] | Jang IC, Oh SJ, Seo JS, et al. Expression of a bifunctional fusion of the Escherichia coli genes for trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase in transgenic rice plants increases trehalose accumulation and abiotic stress tolerance without stunting growth[J]. Plant Physiol, 2003, 131(2): 516-524. |
| [47] |
Romero C, Bellés JM, Vayá JL, et al. Expression of the yeast trehalose-6-phosphate synthase gene in transgenic tobacco plants: pleiotropic phenotypes include drought tolerance[J]. Planta, 1997, 201(3): 293-297.
doi: 10.1007/s004250050069 pmid: 19343407 |
| [48] |
Vishal B, Krishnamurthy P, Ramamoorthy R, et al. OsTPS8 controls yield-related traits and confers salt stress tolerance in rice by enhancing suberin deposition[J]. New Phytol, 2019, 221(3): 1369-1386.
doi: 10.1111/nph.15464 pmid: 30289560 |
| [49] | Veyres N, Danon A, Aono M, et al. The Arabidopsis sweetie mutant is affected in carbohydrate metabolism and defective in the control of growth, development and senescence[J]. Plant J, 2008, 55(4): 665-686. |
| [50] |
Fernandez O, Béthencourt L, Quero A, et al. Trehalose and plant stress responses: friend or foe?[J]. Trends Plant Sci, 2010, 15(7): 409-417.
doi: 10.1016/j.tplants.2010.04.004 pmid: 20494608 |
| [51] | Lunn JE, Feil R, Hendriks JHM, et al. Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucosepyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana[J]. Biochem J, 2006, 397(1): 139-148. |
| [52] |
Nunes C, O'Hara LE, Primavesi LF, et al. The trehalose 6-phosphate/SnRK1 signaling pathway primes growth recovery following relief of sink limitation[J]. Plant Physiol, 2013, 162(3): 1720-1732.
doi: 10.1104/pp.113.220657 pmid: 23735508 |
| [1] | 李勇慧, 鲍星星, 段一珂, 赵运霞, 于相丽, 陈尧, 张延召. 灵宝杜鹃bZIP家族全基因组鉴定及表达特征分析[J]. 生物技术通报, 2024, 40(8): 186-198. |
| [2] | 崔原瑗, 王昭懿, 白双宇, 任毓昭, 豆飞飞, 刘彩霞, 刘凤楼, 王掌军, 李清峰. 大麦非特异性磷脂酶C基因家族全基因组鉴定及苗期胁迫表达分析[J]. 生物技术通报, 2024, 40(8): 74-82. |
| [3] | 余纽, 柳帆, 杨锦昌. 油楠SgTPS7的克隆及其在萜类生物合成和非生物胁迫中的功能[J]. 生物技术通报, 2024, 40(8): 164-173. |
| [4] | 吴丁洁, 陈盈盈, 徐静, 刘源, 张航, 李瑞丽. 植物赤霉素氧化酶及其功能研究进展[J]. 生物技术通报, 2024, 40(7): 43-54. |
| [5] | 王健, 杨莎, 孙庆文, 陈宏宇, 杨涛, 黄园. 金钗石斛bHLH转录因子家族全基因组鉴定及表达分析[J]. 生物技术通报, 2024, 40(6): 203-218. |
| [6] | 胡雅丹, 伍国强, 刘晨, 魏明. MYB转录因子在调控植物响应逆境胁迫中的作用[J]. 生物技术通报, 2024, 40(6): 5-22. |
| [7] | 常雪瑞, 王田田, 王静. 辣椒E2基因家族的鉴定及分析[J]. 生物技术通报, 2024, 40(6): 238-250. |
| [8] | 李景艳, 周家婧, 袁媛, 苏晓艺, 乔文慧, 薛岩磊, 李国婧, 王瑞刚. 拟南芥AtiPGAM2基因参与非生物胁迫的响应[J]. 生物技术通报, 2024, 40(5): 215-224. |
| [9] | 杜兵帅, 邹昕蕙, 王子豪, 张馨元, 曹一博, 张凌云. 油茶SWEET基因家族的全基因组鉴定及表达分析[J]. 生物技术通报, 2024, 40(5): 179-190. |
| [10] | 郭慧妍, 董雪, 安梦楠, 夏子豪, 吴元华. 泛素化修饰关键酶在植物抗逆反应中的功能研究进展[J]. 生物技术通报, 2024, 40(4): 1-11. |
| [11] | 江林琪, 赵佳莹, 郑飞雄, 姚馨怡, 李效贤, 俞振明. 铁皮石斛14-3-3基因家族鉴定及表达分析[J]. 生物技术通报, 2024, 40(3): 229-241. |
| [12] | 周宏丹, 罗晓萍, 涂米雪, 李忠光. 植物褪黑素:植物应答非生物胁迫的新兴信号分子[J]. 生物技术通报, 2024, 40(3): 41-51. |
| [13] | 吴翠翠, 肖水平. 陆地棉HD-Zip家族全基因组鉴定及响应非生物胁迫的表达分析[J]. 生物技术通报, 2024, 40(2): 130-145. |
| [14] | 辛奇, 李压凡, 尹铮, 张晓丹, 陈霆, 刘晓华. 甘蔗CBL-CIPK基因家族的鉴定和表达分析[J]. 生物技术通报, 2024, 40(2): 197-211. |
| [15] | 杨雨青, 谭娟, 汪芳, 彭顺利, 陈婕, 谭明燕, 吕美艳, 周富裕, 刘声传. 茶树叶绿体基因组的研究与应用进展[J]. 生物技术通报, 2024, 40(2): 20-30. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||