








生物技术通报 ›› 2024, Vol. 40 ›› Issue (2): 197-211.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0830
收稿日期:2023-08-25
出版日期:2024-02-26
发布日期:2024-03-13
通讯作者:
刘晓华,女,博士,讲师,研究方向:园艺植物观赏性状与抗逆机理;E-mail: 3305@ldu.edu.cn作者简介:辛奇,男,硕士,研究方向:园艺植物耐逆机理;E-mail: 704427611@qq.com
基金资助:
XIN Qi( ), LI Ya-fan, YIN Zheng, ZHANG Xiao-dan, CHEN Ting, LIU Xiao-hua(
), LI Ya-fan, YIN Zheng, ZHANG Xiao-dan, CHEN Ting, LIU Xiao-hua( )
)
Received:2023-08-25
Published:2024-02-26
Online:2024-03-13
摘要:
【目的】甘蔗是重要的糖料作物,温度、盐碱、水分等因素是制约其生长发育的关键环境因素。类钙调磷酸酶B蛋白CBL(calcineurin B-like protein)是一类Ca2+结合蛋白,通过与其特定的蛋白激酶CIPK(CBL-interacting protein kinase)作用,在Ca2+信号传导通路,尤其是逆境信号传导通路中发挥重要作用。目前,甘蔗全基因组测序已完成,但其CBL-CIPK基因家族成员尚未确定,互作调控机理依然未知。本研究确定了甘蔗CBL、CIPK成员并揭示了CBL-CIPK互作关系,为研究甘蔗CBL-CIPK的互作机理提供基因资源和理论基础。【方法】以甘蔗品种‘GT58’为材料,通过RT-qPCR技术分析CBLs、CIPKs在低温、高温、NaHCO3和PEG处理等4种非生物胁迫下的表达水平,利用酵母双杂交试验分析SsCBLs和SsCIPKs之间的相互作用。【结果】甘蔗全基因组中共有19个CBL基因和82个CIPK基因,分布在不同的进化分支且存在基因复制现象,基因家族成员之间理化性质差异较大,结构域与蛋白基序具有高度保守性,顺式作用元件分布多样;转录水平上,SsCBL7/SsCBL12/SsCIPK1/SsCIPK5的表达水平易受低温、高温、干旱和高盐等多种非生物胁迫的调控;蛋白水平上,SsCBL1与SsCIPK47和SsCIPK81相互作用,SsCBL8与SsCIPK47和SsCIPK81相互作用。【结论】CBL-CIPK互作网络可能在甘蔗生长发育过程中响应非生物胁迫,发挥重要作用。
辛奇, 李压凡, 尹铮, 张晓丹, 陈霆, 刘晓华. 甘蔗CBL-CIPK基因家族的鉴定和表达分析[J]. 生物技术通报, 2024, 40(2): 197-211.
XIN Qi, LI Ya-fan, YIN Zheng, ZHANG Xiao-dan, CHEN Ting, LIU Xiao-hua. Identification and Expression Analysis of the CBL-CIPK Gene Family in Sugarcane[J]. Biotechnology Bulletin, 2024, 40(2): 197-211.
| 基因 Gene | 基因ID Gene ID | 基因长度 Gene length /bp | 正负链 Strand | mRNA长度 mRNA length/bp | 氨基酸数目 Amino acid/aa | 等电点 pI | 外显子个数 Exon number | 染色体定位 Chromosome | 相对分子质量 Molecular weight/kD | 亚细胞定位预测 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|---|
| SsCBL1 | Sspon.01G0014730-1A | 3 124 | + | 579 | 193 | 5.00 | 7 | Chr.1A | 22.26 | Cyto |
| SsCBL2 | Sspon.01G0023690-1A | 3 641 | + | 675 | 225 | 4.82 | 8 | Chr.1A | 25.77 | Cyto |
| SsCBL3 | Sspon.01G0014730-2B | 3 431 | - | 639 | 213 | 4.75 | 8 | Chr.1B | 24.49 | Cyto |
| SsCBL4 | Sspon.01G0014730-3C | 3 431 | - | 639 | 213 | 4.75 | 8 | Chr.1C | 24.49 | Cyto |
| SsCBL5 | Sspon.01G0014730-4D | 3 431 | - | 639 | 213 | 4.75 | 8 | Chr.1D | 24.49 | Cyto |
| SsCBL6 | Sspon.01G0023690-3D | 3 641 | + | 675 | 225 | 4.82 | 8 | Chr.1D | 25.77 | Cyto |
| SsCBL7 | Sspon.02G0030420-1A | 3 582 | + | 660 | 220 | 4.92 | 9 | Chr.2A | 25.43 | Chlo |
| SsCBL8 | Sspon.02G0030420-2C | 3 640 | - | 675 | 225 | 4.79 | 8 | Chr.2C | 25.84 | Cyto |
| SsCBL9 | Sspon.03G0011980-1A | 4 077 | - | 738 | 246 | 6.14 | 10 | Chr.3A | 28.83 | Cyto |
| SsCBL10 | Sspon.03G0012700-1A | 4 617 | - | 882 | 294 | 4.84 | 9 | Chr.3A | 33.11 | E.R. |
| SsCBL11 | Sspon.03G0031480-1B | 4 573 | - | 864 | 288 | 4.84 | 8 | Chr.3B | 32.40 | Chlo |
| SsCBL12 | Sspon.03G0011980-2B | 4 758 | + | 882 | 294 | 5.28 | 10 | Chr.3B | 33.94 | Cyto |
| SsCBL13 | Sspon.03G0011980-1P | 3 620 | - | 660 | 220 | 6.19 | 8 | Chr.3B | 25.66 | Cyto |
| SsCBL14 | Sspon.04G0013450-2B | 3 432 | + | 639 | 213 | 4.77 | 8 | Chr.4B | 24.34 | Nucl |
| SsCBL15 | Sspon.07G0003410-1A | 3 363 | - | 633 | 211 | 4.88 | 8 | Chr.7A | 24.11 | Chlo |
| SsCBL16 | Sspon.07G0018960-1A | 3 619 | - | 669 | 223 | 4.82 | 8 | Chr.7A | 25.68 | Chlo |
| SsCBL17 | Sspon.07G0018960-2B | 3 363 | - | 618 | 206 | 4.99 | 7 | Chr.7B | 23.77 | Chlo |
| SsCBL18 | Sspon.07G0018960-3C | 3 324 | + | 612 | 204 | 4.91 | 7 | Chr.7C | 23.52 | Mito |
| SsCBL19 | Sspon.07G0018960-4D | 3 485 | + | 642 | 214 | 4.92 | 8 | Chr.7D | 24.69 | Chlo |
表1 甘蔗19个CBLs的理化性质统计表
Table 1 Statistical table of physico-chemical properties of 19 CBLs of sugarcane
| 基因 Gene | 基因ID Gene ID | 基因长度 Gene length /bp | 正负链 Strand | mRNA长度 mRNA length/bp | 氨基酸数目 Amino acid/aa | 等电点 pI | 外显子个数 Exon number | 染色体定位 Chromosome | 相对分子质量 Molecular weight/kD | 亚细胞定位预测 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|---|
| SsCBL1 | Sspon.01G0014730-1A | 3 124 | + | 579 | 193 | 5.00 | 7 | Chr.1A | 22.26 | Cyto |
| SsCBL2 | Sspon.01G0023690-1A | 3 641 | + | 675 | 225 | 4.82 | 8 | Chr.1A | 25.77 | Cyto |
| SsCBL3 | Sspon.01G0014730-2B | 3 431 | - | 639 | 213 | 4.75 | 8 | Chr.1B | 24.49 | Cyto |
| SsCBL4 | Sspon.01G0014730-3C | 3 431 | - | 639 | 213 | 4.75 | 8 | Chr.1C | 24.49 | Cyto |
| SsCBL5 | Sspon.01G0014730-4D | 3 431 | - | 639 | 213 | 4.75 | 8 | Chr.1D | 24.49 | Cyto |
| SsCBL6 | Sspon.01G0023690-3D | 3 641 | + | 675 | 225 | 4.82 | 8 | Chr.1D | 25.77 | Cyto |
| SsCBL7 | Sspon.02G0030420-1A | 3 582 | + | 660 | 220 | 4.92 | 9 | Chr.2A | 25.43 | Chlo |
| SsCBL8 | Sspon.02G0030420-2C | 3 640 | - | 675 | 225 | 4.79 | 8 | Chr.2C | 25.84 | Cyto |
| SsCBL9 | Sspon.03G0011980-1A | 4 077 | - | 738 | 246 | 6.14 | 10 | Chr.3A | 28.83 | Cyto |
| SsCBL10 | Sspon.03G0012700-1A | 4 617 | - | 882 | 294 | 4.84 | 9 | Chr.3A | 33.11 | E.R. |
| SsCBL11 | Sspon.03G0031480-1B | 4 573 | - | 864 | 288 | 4.84 | 8 | Chr.3B | 32.40 | Chlo |
| SsCBL12 | Sspon.03G0011980-2B | 4 758 | + | 882 | 294 | 5.28 | 10 | Chr.3B | 33.94 | Cyto |
| SsCBL13 | Sspon.03G0011980-1P | 3 620 | - | 660 | 220 | 6.19 | 8 | Chr.3B | 25.66 | Cyto |
| SsCBL14 | Sspon.04G0013450-2B | 3 432 | + | 639 | 213 | 4.77 | 8 | Chr.4B | 24.34 | Nucl |
| SsCBL15 | Sspon.07G0003410-1A | 3 363 | - | 633 | 211 | 4.88 | 8 | Chr.7A | 24.11 | Chlo |
| SsCBL16 | Sspon.07G0018960-1A | 3 619 | - | 669 | 223 | 4.82 | 8 | Chr.7A | 25.68 | Chlo |
| SsCBL17 | Sspon.07G0018960-2B | 3 363 | - | 618 | 206 | 4.99 | 7 | Chr.7B | 23.77 | Chlo |
| SsCBL18 | Sspon.07G0018960-3C | 3 324 | + | 612 | 204 | 4.91 | 7 | Chr.7C | 23.52 | Mito |
| SsCBL19 | Sspon.07G0018960-4D | 3 485 | + | 642 | 214 | 4.92 | 8 | Chr.7D | 24.69 | Chlo |

图1 CBL-CIPK基因家族的domain与motif分析 A:甘蔗CBL家族成员蛋白基序motif、保守结构域domain分析;B:甘蔗CIPK家族成员motif、domain分析。从左至右分别为进化关系树、motif、domain,不同颜色的模块分别代表不同的motif和domain
Fig. 1 Domain and motif analysis of CBL-CIPK gene family A: Analysis of motif, domain of sugarcane CBL family members. B: Analysis of motif, domain of sugarcane CIPK family members. From left to right are the evolutionary relationship tree, motif and domain, with different colored modules representing different motifs and domains

图2 CBL-CIPK基因家族的基因结构分析 A:甘蔗CBL家族成员基因结构;B:甘蔗CIPK家族成员基因结构;左侧为进化关系树,右侧为家族成员基因结构,绿色部分为UTR,黄色部分为CDS
Fig. 2 Analysis in the gene structures of CBL-CIPK gene family A: Gene structure of sugarcane CBL family members. B: Gene structure of sugarcane CIPK family members. Evolutionary relationship tree on the left, gene structure of family members on the right, UTR in green and CDS in yellow
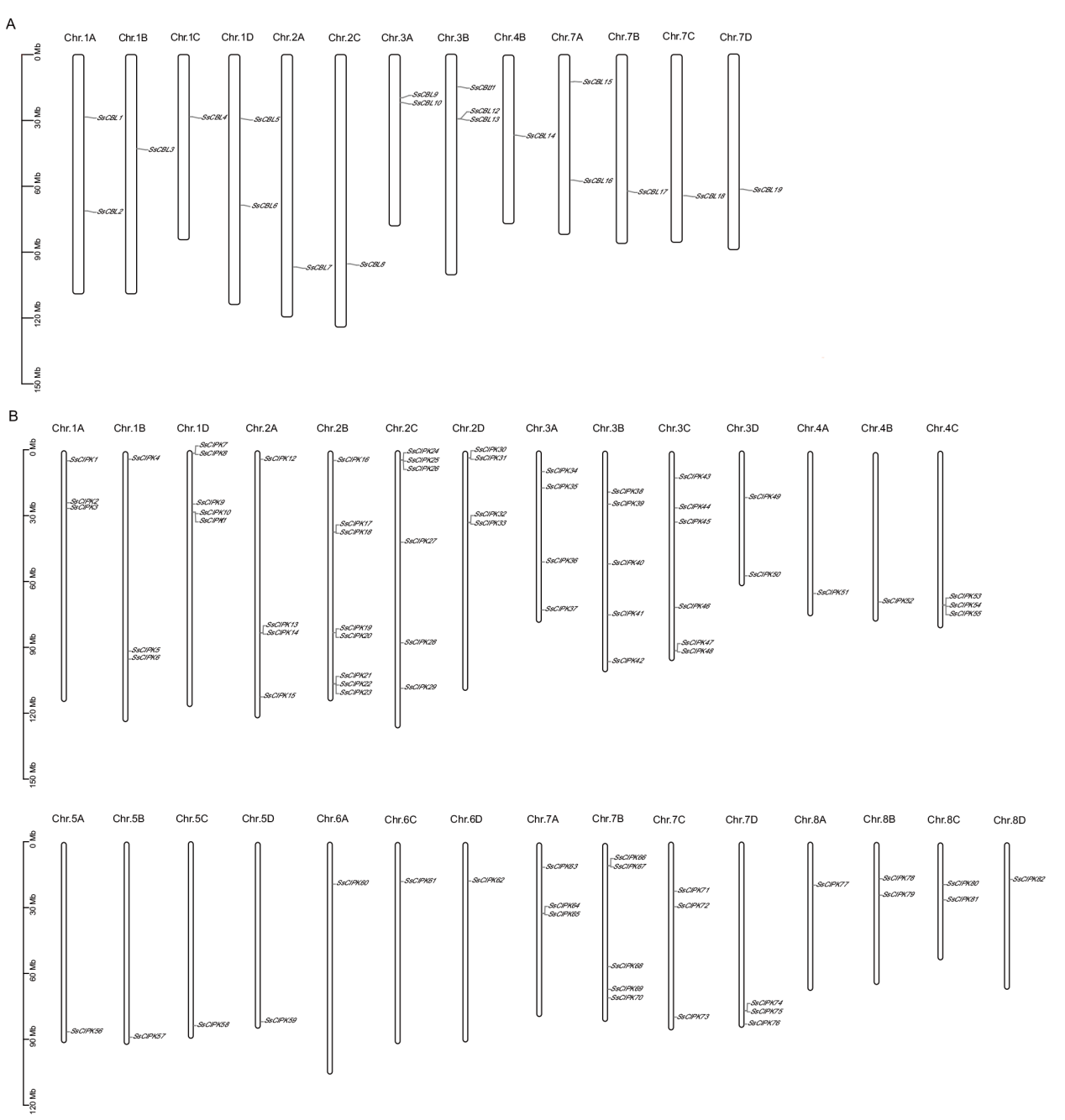
图3 CBL-CIPK基因家族的染色体定位分析 A:甘蔗CBL家族成员染色体定位;B:甘蔗CIPK家族成员染色体定位
Fig. 3 Analysis for their positions on the chromosomes in CBL-CIPK gene family A: Chromosomal localization of sugarcane CBL family members. B: Chromosomal localization of sugarcane CIPK family members
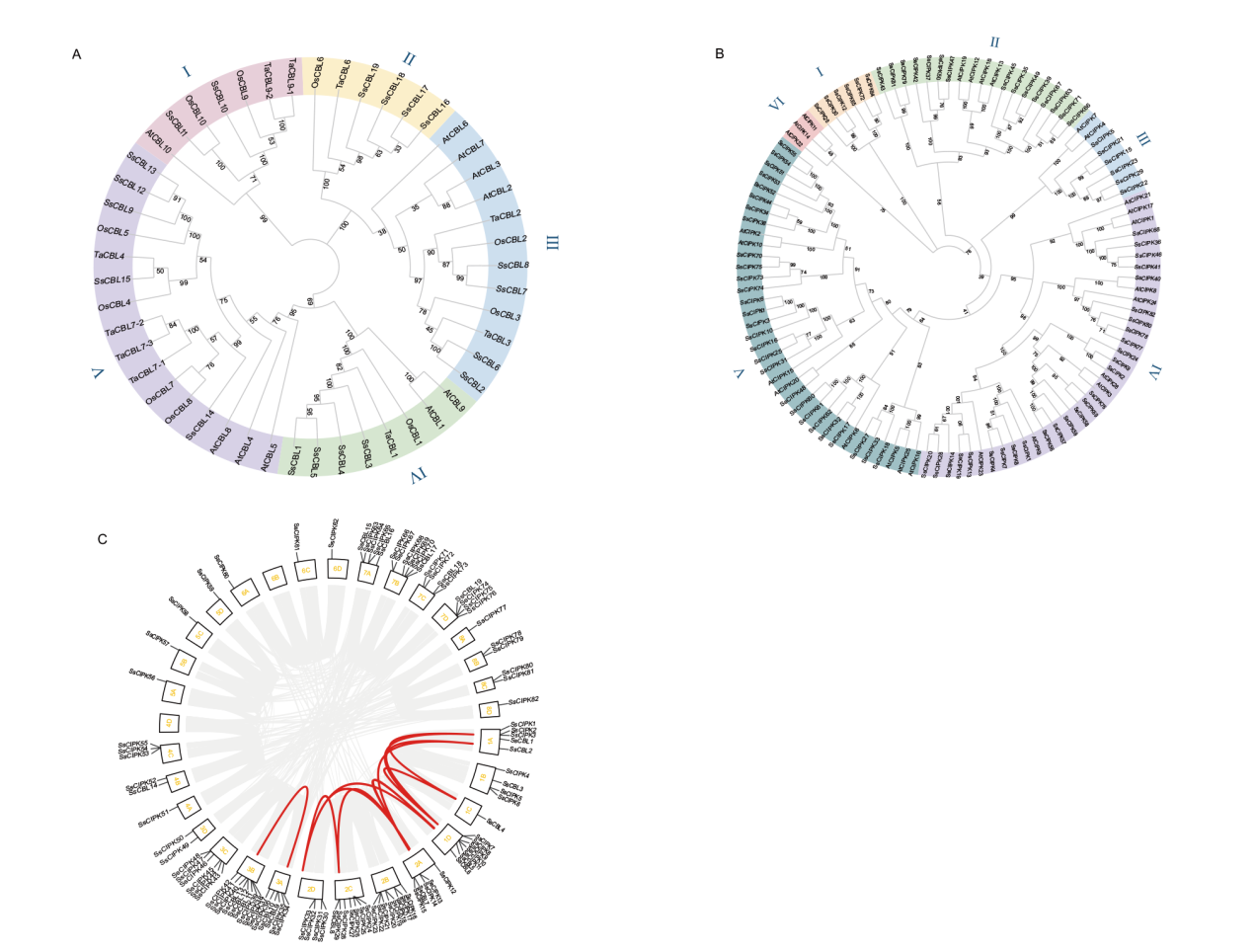
图5 CBL-CIPK基因家族系统发育树及共线性分析 A:多物种CBL家族系统发育进化树;B:多物种CIPK家族系统发育进化树;C:甘蔗种内CBL-CIPK基因家族共线性分析;全基因组中所有基因对均为灰色,CBL-CIPK家族成员形成基因对为红色,甘蔗为异源四倍体,1A-8D为甘蔗的32条染色体
Fig. 5 Phylogenetic tree and covariance analysis of the CBL-CIPK gene family A: Phylogenetic evolutionary tree of the multispecies CBL family. B: Phylogenetic evolutionary tree of the multispecies CIPK family. C: Analysis of intraspecific CBL-CIPK gene family covariance in sugarcane. All gene pairs in the whole genome are gray, CBL-CIPK family members form gene pairs in red, sugarcane is a heterotetraploid, and 1A-8D are the 32 chromosomes of sugarcane

图6 参与不同温度胁迫响应的SsCBLs、SsCIPKs基因的RT-qPCR分析 A:4℃处理下SsCBLs的基因表达谱;B:4℃处理下SsCIPKs的基因表达谱;C:45℃处理下SsCBLs的基因表达谱;D:45℃处理下SsCIPKs的基因表达谱
Fig. 6 RT-qPCR analysis of SsCBLs, SsCIPKs genes involved in different temperature stresses response A: Gene expression profiles of SsCBLs under 4℃ treatment. B: Gene expression profiles of SsCIPKs under 4℃ treatment. C: Gene expression profiles of SsCBLs under 45℃ treatment. D: Gene expression profiles of SsCIPKs under 45℃ treatment

图7 参与NaHCO3/PEG胁迫的SsCBLs、SsCIPKs的RT-qPCR分析 A:170 mmol/L NaHCO3处理下SsCBLs的基因表达谱;B:170 mmol/L NaHCO3处理下SsCIPKs的基因表达谱;C:150 g/L PEG-6000处理下SsCBLs的基因表达谱;D:150 g/L PEG-6000处理下SsCIPKs的基因表达谱
Fig. 7 RT-qPCR analysis of SsCBLs, SsCIPKs genes involved in NaHCO3/PEG stresses A: Gene expression profiles of SsCBLs under170 mmol/L NaHCO3 treatment. B: Gene expression profiles of SsCIPKs under 170 mmol/L NaHCO3 treatment. C: Gene expression profiles of SsCBLs under 150 g/L PEG-6000 treatment. D: Gene expression profiles of SsCIPKs under 150 g/L PEG-6000 treatment
| [1] |
Plasencia FA, Estrada Y, Flores FB, et al. The Ca2+ sensor calcineurin B-like protein 10 in plants: Emerging new crucial roles for plant abiotic stress tolerance[J]. Front Plant Sci, 2021, 11: 599944.
doi: 10.3389/fpls.2020.599944 URL |
| [2] | Niu L, Dong B, Song Z, et al. Genome-wide identification and characterization of CIPK family and analysis responses to various stresses in apple(Malus domestica)[J]. Int J Mol Sci, 2018, 19(7): E2131. |
| [3] |
Hashimoto K, Eckert C, Anschütz U, et al. Phosphorylation of calcineurin B-like(CBL)calcium sensor proteins by their CBL-interacting protein kinases(CIPKs)is required for full activity of CBL-CIPK complexes toward their target proteins[J]. J Biol Chem, 2012, 287(11): 7956-7968.
doi: 10.1074/jbc.M111.279331 pmid: 22253446 |
| [4] | Chaves-Sanjuan A, Sanchez-Barrena MJ, Gonzalez-Rubio JM, et al. Structural basis of the regulatory mechanism of the plant CIPK family of protein kinases controlling ion homeostasis and abiotic stress[J]. Proc Natl Acad Sci USA, 2014, 111(42): E4532-E4541. |
| [5] |
Ma X, Li QH, Yu YN, et al. The CBL-CIPK pathway in plant response to stress signals[J]. Int J Mol Sci, 2020, 21(16): 5668.
doi: 10.3390/ijms21165668 URL |
| [6] |
Mo C, Wan S, Xia Y, et al. Expression patterns and identified protein-protein interactions suggest that cassava CBL-CIPK signal networks function in responses to abiotic stresses[J]. Front Plant Sci, 2018, 9: 269.
doi: 10.3389/fpls.2018.00269 pmid: 29552024 |
| [7] |
Sánchez-Barrena M, Martínez-Ripoll M, Albert A. Structural biology of a major signaling network that regulates plant abiotic stress: The CBL-CIPK mediated pathway[J]. Int J Mol Sci, 2013, 14(3): 5734-5749.
doi: 10.3390/ijms14035734 pmid: 23481636 |
| [8] |
Poddar N, Deepika D, Chitkara P, et al. Molecular and expression analysis indicate the role of CBL interacting protein kinases(CIPKs)in abiotic stress signaling and development in chickpea[J]. Sci Rep, 2022, 12(1): 16862.
doi: 10.1038/s41598-022-20750-2 |
| [9] |
Ródenas R, Vert G. Regulation of root nutrient transporters by CIPK23: “one kinase to rule them all”[J]. Plant Cell Physiol, 2021, 62(4): 553-563.
doi: 10.1093/pcp/pcaa156 pmid: 33367898 |
| [10] |
Yin X, Xia Y, Xie Q, et al. The protein kinase complex CBL10-CIPK8-SOS1 functions in Arabidopsis to regulate salt tolerance[J]. J Exp Bot, 2020, 71(6): 1801-1814.
doi: 10.1093/jxb/erz549 URL |
| [11] |
Imtiaz K, Ahmed M, Annum N, et al. AtCIPK16, a CBL-interacting protein kinase gene, confers salinity tolerance in transgenic wheat[J]. Front Plant Sci, 2023, 14: 1127311.
doi: 10.3389/fpls.2023.1127311 URL |
| [12] |
Gu S, Abid M, Bai D, et al. Transcriptome-wide identification and functional characterization of CIPK gene family members in actinidia valvata under salt stress[J]. Int J Mol Sci, 2023, 24(1): 805.
doi: 10.3390/ijms24010805 URL |
| [13] |
Chen L, Wang QQ, Zhou L, et al. Arabidopsis CBL-interacting protein kinase(CIPK6)is involved in plant response to salt/osmotic stress and ABA[J]. Mol Biol Rep, 2013, 40(8): 4759-4767.
doi: 10.1007/s11033-013-2572-9 URL |
| [14] |
Cui XY, Du YT, Fu JD, et al. Wheat CBL-interacting protein kinase 23 positively regulates drought stress and ABA responses[J]. BMC Plant Biol, 2018, 18(1): 93.
doi: 10.1186/s12870-018-1306-5 |
| [15] |
Gao C, Lu S, Zhou R, et al. The OsCBL8-OsCIPK17 module regulates seedling growth and confers resistance to heat and drought in rice[J]. Int J Mol Sci, 2022, 23(20): 12451.
doi: 10.3390/ijms232012451 URL |
| [16] |
Meng D, Dong BY, Niu LL, et al. The pigeon pea CcCIPK14-CcCBL1 pair positively modulates drought tolerance by enhancing flavonoid biosynthesis[J]. Plant J, 2021, 106(5): 1278-1297.
doi: 10.1111/tpj.v106.5 URL |
| [17] |
Yang L, Zhang N, Wang K, et al. CBL-interacting protein kinases 18(CIPK18)gene positively regulates drought resistance in potato[J]. Int J Mol Sci, 2023, 24(4): 3613.
doi: 10.3390/ijms24043613 URL |
| [18] |
Yang S, Li J, Lu J, et al. Potato calcineurin B-like protein CBL4, interacting with calcineurin B-like protein-interacting protein kinase CIPK2, positively regulates plant resistance to stem canker caused by Rhizoctonia solani[J]. Front Microbiol, 2023, 13: 1032900.
doi: 10.3389/fmicb.2022.1032900 URL |
| [19] |
Piao HL, Xuan YH, Park SH, et al. OsCIPK31, a CBL-interacting protein kinase is involved in germination and seedling growth under abiotic stress conditions in rice plants[J]. Mol Cells, 2010, 30(1): 19-27.
doi: 10.1007/s10059-010-0084-1 URL |
| [20] |
Liu P, Duan YH, Liu C, et al. The calcium sensor TaCBL4 and its interacting protein TaCIPK5 are required for wheat resistance to stripe rust fungus[J]. J Exp Bot, 2018, 69(18): 4443-4457.
doi: 10.1093/jxb/ery227 pmid: 29931351 |
| [21] |
Xi Y, Liu JY, Dong C, et al. The CBL and CIPK gene family in grapevine(Vitis vinifera): Genome-wide analysis and expression profiles in response to various abiotic stresses[J]. Front Plant Sci, 2017, 8: 978.
doi: 10.3389/fpls.2017.00978 URL |
| [22] | Aslam M, Fakher B, Jakada BH, et al. Genome-wide identification and expression profiling of CBL-CIPK gene family in pineapple(Ananas comosus)and the role of AcCBL1 in abiotic and biotic stress response[J]. Biomolecules, 2019, 9(7): E293. |
| [23] |
Zhang HF, Yang B, Liu WZ, et al. Identification and characterization of CBL and CIPK gene families in canola(Brassica napus L.)[J]. BMC Plant Biol, 2014, 14(1): 8.
doi: 10.1186/1471-2229-14-8 |
| [24] |
Yin X, Wang QL, Chen Q, et al. Genome-wide identification and functional analysis of the calcineurin B-like protein and calcineurin B-like protein-interacting protein kinase gene families in turnip(Brassica rapa var. rapa)[J]. Front Plant Sci, 2017, 8: 1191.
doi: 10.3389/fpls.2017.01191 URL |
| [25] |
Wang QW, Zhao K, Gong YQ, et al. Genome-wide identification and functional analysis of the calcineurin B-like protein and calcineurin B-like protein-interacting protein kinase gene families in chinese cabbage(Brassica rapa ssp. pekinensis)[J]. Genes, 2022, 13(5): 795.
doi: 10.3390/genes13050795 URL |
| [26] |
Mao JJ, Manik SM, Shi SJ, et al. Mechanisms and physiological roles of the CBL-CIPK networking system in Arabidopsis thaliana[J]. Genes, 2016, 7(9): 62.
doi: 10.3390/genes7090062 URL |
| [27] |
Chen Y, Jin YF, Wang YS, et al. Diverse roles of the CIPK gene family in transcription regulation and various biotic and abiotic stresses: A literature review and bibliometric study[J]. Front Genet, 2022, 13: 1041078.
doi: 10.3389/fgene.2022.1041078 URL |
| [28] |
Zhu XL, Wang BQ, Wang X, et al. Identification of the CIPK-CBL family gene and functional characterization of CqCIPK14 gene under drought stress in quinoa[J]. BMC Genomics, 2022, 23(1): 447.
doi: 10.1186/s12864-022-08683-6 pmid: 35710332 |
| [29] |
Zhang XX, Ren XL, Qi XT, et al. Evolution of the CBL and CIPK gene families in Medicago: genome-wide characterization, pervasive duplication, and expression pattern under salt and drought stress[J]. BMC Plant Biol, 2022, 22(1): 512.
doi: 10.1186/s12870-022-03884-3 |
| [30] |
Tang J, Lin J, Li H, et al. Characterization of CIPK family in Asian pear(Pyrus bretschneideri Rehd)and Co-expression analysis related to salt and osmotic stress responses[J]. Front Plant Sci, 2016, 7: 1361.
doi: 10.3389/fpls.2016.01361 pmid: 27656193 |
| [31] |
Cho JH, Choi MN, Yoon KH, et al. Ectopic expression of sjcbl1, calcineurin b-like 1 gene from sedirea japonica, rescues the salt and osmotic stress hypersensitivity in arabidopsis cbl1 mutant[J]. Front Plant Sci, 2018, 9: 1188.
doi: 10.3389/fpls.2018.01188 URL |
| [32] |
Huang SL, Jiang SF, Liang JS, et al. Roles of plant CBL-CIPK systems in abiotic stress responses[J]. Turk J Bot, 2019, 43(3): 271-280.
doi: 10.3906/bot-1810-35 |
| [33] |
Chao M, Dong J, Hu G, et al. Sequence characteristics and expression analysis of GhCIPK23 gene in upland cotton(Gossypium hirsutum L.)[J]. Int J Mol Sci, 2022, 23(19): 12040.
doi: 10.3390/ijms231912040 URL |
| [34] |
Zhu KK, Chen F, Liu JY, et al. Evolution of an intron-poor cluster of the CIPK gene family and expression in response to drought stress in soybean[J]. Sci Rep, 2016, 6(1): 28225.
doi: 10.1038/srep28225 |
| [35] |
Zhu KK, Fan PH, Liu H, et al. Insight into the CBL and CIPK gene families in pecan(Carya illinoinensis): identification, evolution and expression patterns in drought response[J]. BMC Plant Biol, 2022, 22(1): 221.
doi: 10.1186/s12870-022-03601-0 |
| [36] |
Ma X, Gai WX, Li Y, et al. The CBL-interacting protein kinase CaCIPK13 positively regulates defence mechanisms against cold stress in pepper[J]. J Exp Bot, 2022, 73(5): 1655-1667.
doi: 10.1093/jxb/erab505 pmid: 35137060 |
| [37] |
Xiao C, Zhang H, Xie F, et al. Evolution, gene expression, and protein-protein interaction analyses identify candidate CBL-CIPK signalling networks implicated in stress responses to cold and bacterial infection in citrus[J]. BMC Plant Biol, 2022, 22(1): 420.
doi: 10.1186/s12870-022-03809-0 pmid: 36045357 |
| [38] |
Zhou YL, Cheng Y, Yang YQ, et al. Overexpression of SpCBL6, a calcineurin B-like protein of Stipa purpurea, enhanced cold tolerance and reduced drought tolerance in transgenic Arabidopsis[J]. Mol Biol Rep, 2016, 43(9): 957-966.
doi: 10.1007/s11033-016-4036-5 URL |
| [39] |
Qiu K, Pan H, Sheng Y, et al. The peach(Prunus persica)CBL and CIPK family genes: Protein interaction profiling and expression analysis in response to various abiotic stresses[J]. Plants, 2022, 11(21): 3001.
doi: 10.3390/plants11213001 URL |
| [40] |
Wang S, Li Q. Genome-wide identification of the Salvia miltiorrhiza SmCIPK gene family and revealing the salt resistance characteristic of SmCIPK13[J]. Int J Mol Sci, 2022, 23(12): 6861.
doi: 10.3390/ijms23126861 URL |
| [41] |
Huang L, Li Z, Fu Q, et al. Genome-wide identification of CBL-CIPK gene family in honeysuckle(Lonicera japonica Thunb.)and their regulated expression under salt stress[J]. Front Genet, 2021, 12: 751040.
doi: 10.3389/fgene.2021.751040 URL |
| [42] |
Ma X, Gai WX, Qiao YM, et al. Identification of CBL and CIPK gene families and functional characterization of CaCIPK1 under Phytophthora capsici in pepper(Capsicum annuum L.)[J]. BMC Genomics, 2019, 20(1): 775.
doi: 10.1186/s12864-019-6125-z |
| [43] |
Ma R, Liu W, Li S, et al. Genome-wide identification, characterization and expression analysis of the CIPK gene family in potato(Solanum tuberosum L.)and the role of StCIPK10 in response to drought and osmotic stress[J]. Int J Mol Sci, 2021, 22(24): 13535.
doi: 10.3390/ijms222413535 URL |
| [44] |
Wang L, Feng X, Yao L, et al. Characterization of CBL-CIPK signaling complexes and their involvement in cold response in tea plant[J]. Plant Physiol Biochem, 2020, 154: 195-203.
doi: 10.1016/j.plaphy.2020.06.005 URL |
| [45] |
Sun W, Zhang B, Deng J, et al. Genome-wide analysis of CBL and CIPK family genes in cotton: conserved structures with divergent interactions and expression[J]. Physiol Mol Biol Plants, 2021, 27(2): 359-368.
doi: 10.1007/s12298-021-00943-1 |
| [1] | 吴翠翠, 肖水平. 陆地棉HD-Zip家族全基因组鉴定及响应非生物胁迫的表达分析[J]. 生物技术通报, 2024, 40(2): 130-145. |
| [2] | 任延靖, 张鲁刚, 赵孟良, 李江, 邵登魁. 白菜种子cDNA酵母文库的构建及BrTTG1互作蛋白的筛选及分析[J]. 生物技术通报, 2024, 40(2): 223-232. |
| [3] | 邹修为, 岳佳妮, 李志宇, 戴良英, 李魏. 水稻热激转录因子HsfA2b调控非生物胁迫抗性的功能分析[J]. 生物技术通报, 2024, 40(2): 90-98. |
| [4] | 张怡, 张心如, 张金珂, 胡利宗, 上官欣欣, 郑晓红, 胡娟娟, 张聪聪, 穆桂清, 李成伟. 小麦镉胁迫响应基因TaMYB1的功能分析[J]. 生物技术通报, 2024, 40(1): 194-206. |
| [5] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [6] | 温晓蕾, 李建嫄, 李娜, 张娜, 杨文香. 小麦叶锈菌与小麦互作的酵母双杂交cDNA文库构建与应用[J]. 生物技术通报, 2023, 39(9): 136-146. |
| [7] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [8] | 李苑虹, 郭昱昊, 曹燕, 祝振洲, 王飞飞. 外源植物激素调控微藻生长及目标产物积累研究进展[J]. 生物技术通报, 2023, 39(6): 61-72. |
| [9] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [10] | 李心怡, 姜春秀, 薛丽, 蒋洪涛, 姚伟, 邓祖湖, 张木清, 余凡. 多荧光标记引物增强甘蔗染色体寡聚核苷酸探针杂交信号[J]. 生物技术通报, 2023, 39(5): 103-111. |
| [11] | 翟莹, 李铭杨, 张军, 赵旭, 于海伟, 李珊珊, 赵艳, 张梅娟, 孙天国. 异源表达大豆转录因子GmNF-YA19提高转基因烟草抗旱性[J]. 生物技术通报, 2023, 39(5): 224-232. |
| [12] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [13] | 侯筱媛, 车郑郑, 李姮静, 杜崇玉, 胥倩, 王群青. 大豆膜系统cDNA文库的构建及大豆疫霉效应子PsAvr3a互作蛋白的筛选[J]. 生物技术通报, 2023, 39(4): 268-276. |
| [14] | 杨春洪, 董璐, 陈林, 宋丽. 大豆VAS1基因家族的鉴定及参与侧根发育的研究[J]. 生物技术通报, 2023, 39(3): 133-142. |
| [15] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||