








生物技术通报 ›› 2024, Vol. 40 ›› Issue (8): 221-231.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0112
李亦君1,2( ), 杨小贝2, 夏琳2, 罗朝鹏2, 徐馨2, 杨军2, 宁黔冀1, 武明珠2(
), 杨小贝2, 夏琳2, 罗朝鹏2, 徐馨2, 杨军2, 宁黔冀1, 武明珠2( )
)
收稿日期:2024-01-31
出版日期:2024-08-26
发布日期:2024-07-02
通讯作者:
武明珠,女,博士,高级工程师,研究方向:烟草功能基因;E-mail: mingzhuwus@126.com作者简介:李亦君,女,硕士,研究方向:烟草功能基因;E-mail: liyijun124@163.com
基金资助:
LI Yi-jun1,2( ), YANG Xiao-bei2, XIA Lin2, LUO Zhao-peng2, XU Xin2, YANG Jun2, NING Qian-ji1, WU Ming-zhu2(
), YANG Xiao-bei2, XIA Lin2, LUO Zhao-peng2, XU Xin2, YANG Jun2, NING Qian-ji1, WU Ming-zhu2( )
)
Received:2024-01-31
Published:2024-08-26
Online:2024-07-02
摘要:
【目的】伪答应基因家族(pseudo response regulators, PRRs)是高等植物调控开花途径的重要基因。克隆烟草NtPRR37基因并分析其对不同光周期的应答及对开花的影响,为烟草开花调控提供靶标基因。【方法】利用同源克隆方法从普通烟草(Nicotiana tabacum L.)中克隆得到NtPRR37基因,并对其进行生物信息学分析,利用实时荧光定量PCR(real-time quantitative PCR, RT-qPCR)分析其在不同组织中的表达及不同光照时长处理的表达模式。同时利用病毒诱导基因沉默(virus induced gene silence, VIGS)技术降低NtPRR37表达水平并观察表型变化及检测开花相关基因表达变化。【结果】 NtPRR37基因全长2 472 bp,编码823个氨基酸,相对分子质量90.16 kD,含有PRRs基因家族的典型保守结构域(REC和CCT结构域)。通过同源进化分析发现,烟草NtPRR37与绒毛烟草(Nicotiana tomentosiformis)、林烟草(Nicotiana sylvestris)及本氏烟草(Nicotiana benthamiana)的PRR37在进化上属于同一分支。采用RT-qPCR分析发现,该基因在盛花期烟草各个组织的表达特征存在差异性,在雌蕊中的表达量最高,在侧根的表达量最低;在不同光照时长处理下,NtPRR37随着光照时间的增加表达量呈上升趋势,全黑暗处理下表达量最低,且具有生物节律性;NtPRR37沉默植株中NtPRR37表达量明显下调且沉默植株开花期提前,这可能与诱导开花相关基因(NtFT4、NtAP1、NtCO、NtSOC1)表达量显著上调有关。【结论】NtPRR37的表达受到光周期的调控,且在烟草开花过程中NtPRR37作为开花抑制因子存在。
李亦君, 杨小贝, 夏琳, 罗朝鹏, 徐馨, 杨军, 宁黔冀, 武明珠. 烟草NtPRR37基因克隆及功能分析[J]. 生物技术通报, 2024, 40(8): 221-231.
LI Yi-jun, YANG Xiao-bei, XIA Lin, LUO Zhao-peng, XU Xin, YANG Jun, NING Qian-ji, WU Ming-zhu. Cloning and Functional Analysis of NtPRR37 Gene in Nicotiana tabacum L.[J]. Biotechnology Bulletin, 2024, 40(8): 221-231.
| 引物名称Primer name | 碱基序列Base sequence(5'-3') | 用途Usage |
|---|---|---|
| NtPRR37-F | GAGGAAGATGAGTCAAGGAT | 基因克隆 Cloning of gene |
| NtPRR37-R | TCTGTTCTGCGAGTCTCT | |
| qNtPRR37-F | ACCATCATCACTACCATCAC | NtPRR37基因的RT-qPCR 检测 RT-qPCR detection of NtPRR37 gene |
| qNtPRR37-R | TGCTTCCATTGTTACTTCCT | |
| L25-F | CCCCTCACCACAGAGTCTGC | 内参基因的RT-qPCR检测 RT-qPCR detection of internal reference gene |
| L25-R | AAGGGTGTTGTTGTCCTCAATCTT | |
| NtPRR37-VIGS-F | CACTTGTGCCCAGGTTGTC | VIGS载体的构建 Construction of VIGS vector |
| NtPRR37-VIGS-R | TCTAGGAGCGGCTACATCGT | |
| NtFT4-F | GTCACAGACATCCCAGCAACT | NtFT4基因的RT-qPCR检测 RT-qPCR detection of NtFT4 gene |
| NtFT4-R | CGAAACACTACGAAAACAAAGC | |
| NtAP1-F | CCTTACACCTTTTCTCAGACCAA | NtAP1基因的RT-qPCR检测 RT-qPCR detection of NtAP1gene |
| NtAP1-R | ATGTGCTTTCTTCGCTAAACCTC | |
| NtCO-F | CAAATATGGCTCCTCAGGGA | NtCO基因的RT-qPCR检测 RT-qPCR detection of NtCO gene |
| NtCO-R | GGATGAAATGTATGCGTTATGG | |
| NtSOC1-F | AAACAGTTGGAGCGGAGTG | NtSOC1基因的RT-qPCR检测 RT-qPCR detection of NtSOC1 gene |
| NtSOC1-R | GCATTTTCAGAAGCAAGGAT |
表1 本实验所涉及引物及其名称
Table 1 Primers and their names involved in this experiment
| 引物名称Primer name | 碱基序列Base sequence(5'-3') | 用途Usage |
|---|---|---|
| NtPRR37-F | GAGGAAGATGAGTCAAGGAT | 基因克隆 Cloning of gene |
| NtPRR37-R | TCTGTTCTGCGAGTCTCT | |
| qNtPRR37-F | ACCATCATCACTACCATCAC | NtPRR37基因的RT-qPCR 检测 RT-qPCR detection of NtPRR37 gene |
| qNtPRR37-R | TGCTTCCATTGTTACTTCCT | |
| L25-F | CCCCTCACCACAGAGTCTGC | 内参基因的RT-qPCR检测 RT-qPCR detection of internal reference gene |
| L25-R | AAGGGTGTTGTTGTCCTCAATCTT | |
| NtPRR37-VIGS-F | CACTTGTGCCCAGGTTGTC | VIGS载体的构建 Construction of VIGS vector |
| NtPRR37-VIGS-R | TCTAGGAGCGGCTACATCGT | |
| NtFT4-F | GTCACAGACATCCCAGCAACT | NtFT4基因的RT-qPCR检测 RT-qPCR detection of NtFT4 gene |
| NtFT4-R | CGAAACACTACGAAAACAAAGC | |
| NtAP1-F | CCTTACACCTTTTCTCAGACCAA | NtAP1基因的RT-qPCR检测 RT-qPCR detection of NtAP1gene |
| NtAP1-R | ATGTGCTTTCTTCGCTAAACCTC | |
| NtCO-F | CAAATATGGCTCCTCAGGGA | NtCO基因的RT-qPCR检测 RT-qPCR detection of NtCO gene |
| NtCO-R | GGATGAAATGTATGCGTTATGG | |
| NtSOC1-F | AAACAGTTGGAGCGGAGTG | NtSOC1基因的RT-qPCR检测 RT-qPCR detection of NtSOC1 gene |
| NtSOC1-R | GCATTTTCAGAAGCAAGGAT |
| 名称 Name | 网址 Website | 用途 Usage |
|---|---|---|
| Expasy | | 基因序列翻译成氨基酸序列Gene sequence translated into amino acid sequence |
| ProtParam | | 分析氨基酸含量、分子量和等电点Analysis of amino acid content, molecular weight and isoelectric point |
| Protscale | | 分析蛋白的亲水性及疏水性Analysis of hydrophilicity and hydrophobicity of proteins |
| SMART | | 分析保守结构域Analysis of conserved domains |
| SOPMA | | 预测二级结构Prediction of secondary structure |
| SWISS-MODEL | | 预测三级结构Prediction of tertiary structure |
| SignalP-5.0 | | 分析信号肽Analysis of signal peptide |
表2 生物信息学在线工具
Table 2 Bioinformatics online tools
| 名称 Name | 网址 Website | 用途 Usage |
|---|---|---|
| Expasy | | 基因序列翻译成氨基酸序列Gene sequence translated into amino acid sequence |
| ProtParam | | 分析氨基酸含量、分子量和等电点Analysis of amino acid content, molecular weight and isoelectric point |
| Protscale | | 分析蛋白的亲水性及疏水性Analysis of hydrophilicity and hydrophobicity of proteins |
| SMART | | 分析保守结构域Analysis of conserved domains |
| SOPMA | | 预测二级结构Prediction of secondary structure |
| SWISS-MODEL | | 预测三级结构Prediction of tertiary structure |
| SignalP-5.0 | | 分析信号肽Analysis of signal peptide |
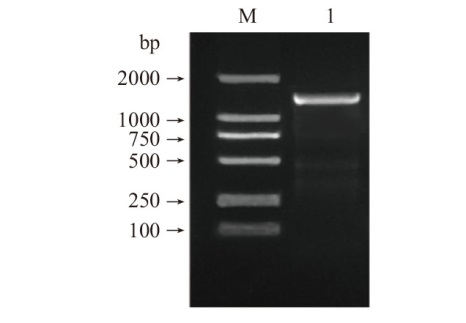
图1 NtPRR37基因的CDS序列PCR产物电泳图 M:Marker 5000;1:NtPRR37基因CDS序列扩增产物
Fig. 1 Electrophoretogram of PCR product of NtPRR37 gene CDS sequence M: Marker 5000. 1: Amplified product of CDS sequence of NtPRR37

图2 烟草与其他物种PRR37氨基酸序列多重比对 图中红色方框内容为PRR37蛋白的结构域(REC和CCT结构域)
Fig. 2 Multiple sequence alignment of PRR37 between tobacco and other species The red box in the diagram contains the domains(REC and CCT domains)of PRR37 protein
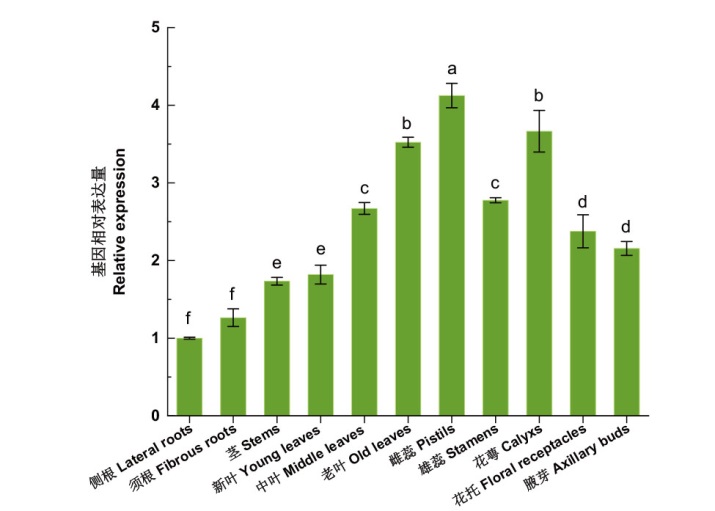
图4 烟草NtPRR37基因在烟草盛花期不同组织中的表达 不同小写字母表示差异达到显著水平(P < 0.05),下同
Fig. 4 Expressions of NtPRR37 gene in different tissues of tobacco in blooming period Different lower letters indicate significant difference(P<0.05)
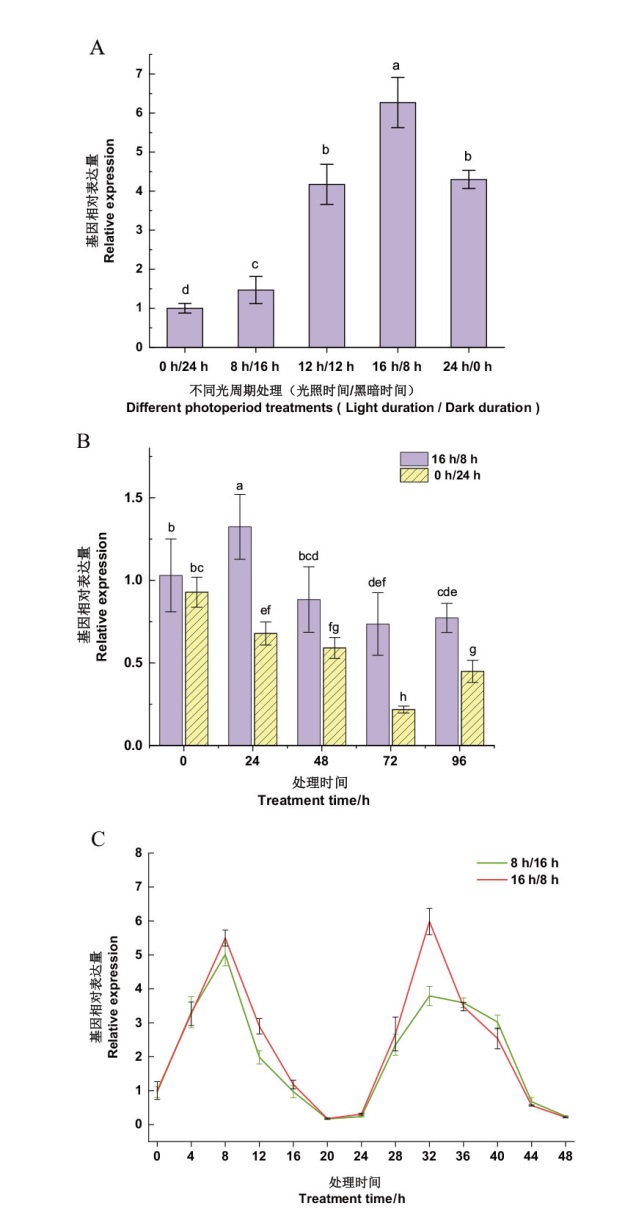
图5 光照对NtPRR37基因表达的影响 A:不同光周期对NtPRR37基因表达的影响;B:不同黑暗时长处理对NtPRR37基因表达的影响(黑暗处理为0 h/24 h,长光照处理为16 h/8 h); C:连续长光照和短光照NtPRR37基因的表达情况分析(短光照处理为8 h/16 h,长光照为16 h/8 h)
Fig. 5 Effects of light on the expression of NtPRR37 gene A: The effect of different photoperiods on the expression of NtPRR37 gene. B: The effect of different darkness durations on the expression of NtPRR37 gene(Darkness treatment: 0 h/24 h, light treatment:16 h/8 h). C : Analysis of the expression of NtPRR37 gene under continuous long light and short light(Short light treatment: 8 h/16 h, long light treatment: 16 h/8 h)
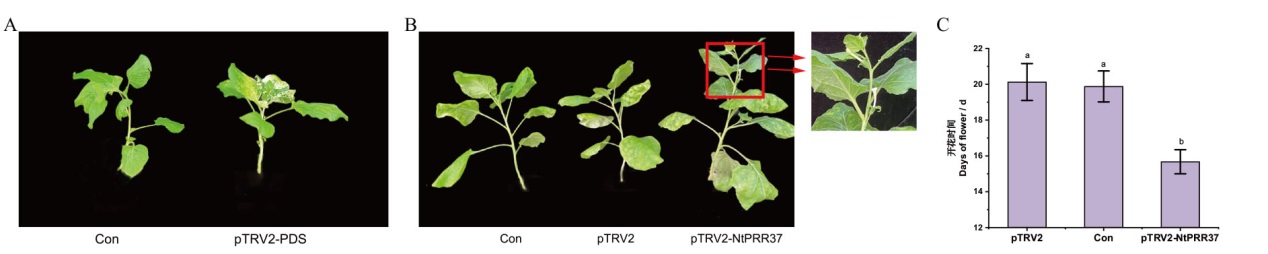
图6 农杆菌侵染本氏烟草后的烟草表型 A:空白对照(Con)与阳性对照(pTRV2-PDS)植株侵染7d后的表型对比;B:阴性对照(pTRV2)、空白对照(Con)及实验组(pTRV2-NtPRR37)植株侵染16 d后的表型对比;C:阴性对照(pTRV2)、空白对照(Con)及实验组(pTRV2-NtPRR37)植株开花时间对比
Fig. 6 Phenotype of Nicotiana benthamiana after Agrobacterium infection A: Phenotypic comparison of blank control(Con)and positive control(pTRV2-PDS)plants after 7 d of infection. B: Phenotypic comparison of negative control(pTRV2), blank control(Con)and experimental group(pTRV2-NtPRR37)plants after 16 d of infection. C: Comparison of flowering time of negative control(pTRV2), blank control(Con)and experimental group(pTRV2-NtPRR37)plants
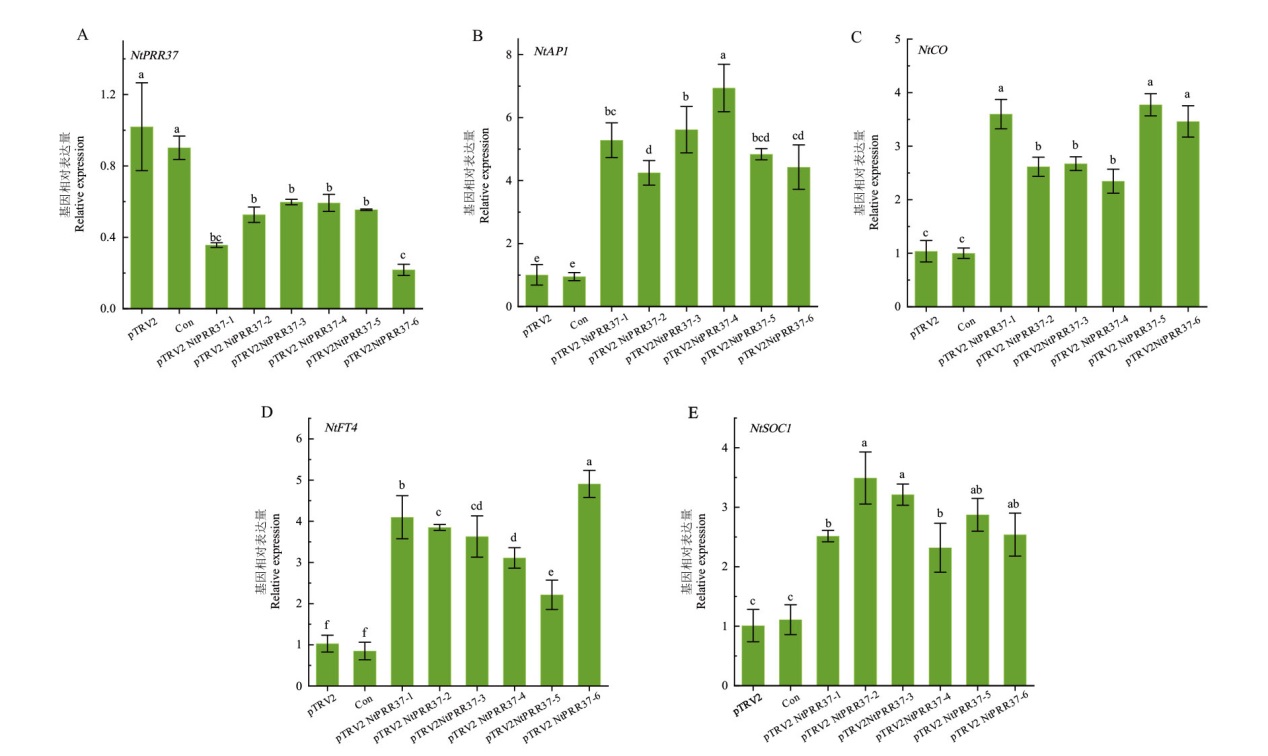
图7 NtPRR37基因沉默后开花相关基因表达分析 阴性对照(pTRV2)、空白对照(Con)及实验组(pTRV2-NtPRR37)植株侵染15 d后的相关基因表达分析
Fig. 7 Expression analysis of genes related to flowering after NtPRR37 gene silenced Expression analysis of related genes after 15 d of infection in negative control (pTRV2), blank control (Con) and experimental group (pTRV2-NtPRR37) plants
| [1] | 杨甲甲, 杨米连, 胡彦如. 生物钟PRR蛋白促进拟南芥幼苗中花青素的合成[J]. 广西植物, 2023, 43(4): 676-687. |
| Yang JJ, Yang ML, Hu YR. Circadian clock PRR proteins stimulate anthocyanin synthesis in Arabidopsis thaliana seedlings[J]. Guihaia, 2023, 43(4): 676-687. | |
| [2] | Hargreaves JK, Oakenfull RJ, Davis AM, et al. Multiple metals influence distinct properties of the Arabidopsis circadian clock[J]. PLoS One, 2022, 17(4): e0258374. |
| [3] | 莫伟亮. 拟南芥蓝光受体CRY2介导相分离调控生物钟机制的研究[D]. 长春: 吉林大学, 2022. |
| Mo WL. Study on the mechanism of Arabidopsis blue light receptor CRY2-mediated phase separation to regulate the circadian clock[D]. Changchun: Jilin University, 2022. | |
| [4] | Mizuno T, Nakamichi N. Pseudo-response regulators(PRRs)or true oscillator components(TOCs)[J]. Plant Cell Physiol, 2005, 46(5): 677-685. |
| [5] | Koo BH, Yoo SC, Park JW, et al. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes[J]. Mol Plant, 2013, 6(6): 1877-1888. |
| [6] | Salomé PA, McClung CR. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock[J]. Plant Cell, 2005, 17(3): 791-803. |
| [7] |
Zhang L, Li QP, Dong HJ, et al. Three CCT domain-containing genes were identified to regulate heading date by candidate gene-based association mapping and transformation in rice[J]. Sci Rep, 2015, 5: 7663.
doi: 10.1038/srep07663 pmid: 25563494 |
| [8] | Cockram J, Thiel T, Steuernagel B, et al. Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae[J]. PLoS One, 2012, 7(9): e45307. |
| [9] | Nimmo HG, Laird J. Arabidopsis thaliana PRR7 provides circadian input to the CCA1 promoter in shoots but not roots[J]. Front Plant Sci, 2021, 12: 750367. |
| [10] | Yuan L, Hu Y, Li SL, et al. PRR9 and PRR7 negatively regulate the expression of EC components under warm temperature in roots[J]. Plant Signal Behav, 2021, 16(2): 1855384. |
| [11] |
He YQ, Yu YJ, Wang XL, et al. Aschoff's rule on circadian rhythms orchestrated by blue light sensor CRY2 and clock component PRR9[J]. Nat Commun, 2022, 13(1): 5869.
doi: 10.1038/s41467-022-33568-3 pmid: 36198686 |
| [12] |
Wang L, Kim J, Somers DE. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription[J]. Proc Natl Acad Sci USA, 2013, 110(2): 761-766.
doi: 10.1073/pnas.1215010110 pmid: 23267111 |
| [13] |
Zhang B, Liu HY, Qi FX, et al. Genetic interactions among Ghd7, Ghd8, OsPRR37 and Hd1 contribute to large variation in heading date in rice[J]. Rice, 2019, 12(1): 48.
doi: 10.1186/s12284-019-0314-x pmid: 31309345 |
| [14] | Klein RR, Miller FR, Dugas DV, et al. Allelic variants in the PRR37 gene and the human-mediated dispersal and diversification of sorghum[J]. Theor Appl Genet, 2015, 128(9): 1669-1683. |
| [15] | Murphy RL, Klein RR, Morishige DT, et al. Coincident light and clock regulation of pseudoresponse regulator protein 37(PRR37)controls photoperiodic flowering in sorghum[J]. Proc Natl Acad Sci USA, 2011, 108(39): 16469-16474. |
| [16] | Kamioka M, Takao SR, Suzuki T, et al. Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock[J]. Plant Cell, 2016, 28(3): 696-711. |
| [17] |
Mizoguchi T, Wright L, Fujiwara S, et al. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis[J]. Plant Cell, 2005, 17(8): 2255-2270.
doi: 10.1105/tpc.105.033464 pmid: 16006578 |
| [18] | Blázquez M. Flower development pathways[J]. J Cell Sci, 2000, 113(Pt 20): 3547-3548. |
| [19] | Komeda Y. Genetic regulation of time to flower in Arabidopsis thaliana[J]. Annu Rev Plant Biol, 2004, 55: 521-535. |
| [20] | Harig L, Beinecke FA, Oltmanns J, et al. Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco[J]. Plant J, 2012, 72(6): 908-921. |
| [21] |
Schmidt GW, Delaney SK. Stable internal reference genes for normalization of real-time RT-PCR in tobacco(Nicotiana tabacum)during development and abiotic stress[J]. Mol Genet Genomics, 2010, 283(3): 233-241.
doi: 10.1007/s00438-010-0511-1 pmid: 20098998 |
| [22] | 武明珠, 刘瑞霞, 王中, 等. 利用VIGS技术研究NtbHLH93基因在烟草甾醇代谢中的功能[J]. 烟草科技, 2019, 52(6): 16-22. |
| Wu MZ, Liu RX, Wang Z, et al. Study on function of NtbHLH93 in sterol metabolism of tobacco by VIGS[J]. Tob Sci Technol, 2019, 52(6): 16-22. | |
| [23] |
李剑峰, 李婷, 贾小平. PRRs家族功能基因的研究进展[J]. 植物遗传资源学报, 2019, 20(6): 1399-1407.
doi: 10.13430/j.cnki.jpgr.20190403001 |
| Li JF, Li T, Jia XP. Advances on unlocking the functional basis of PRRs family genes[J]. J Plant Genet Resour, 2019, 20(6): 1399-1407. | |
| [24] |
Nakamichi N, Takao SR, Kudo T, et al. Improvement of Arabidopsis biomass and cold, drought and salinity stress tolerance by modified circadian clock-associated PSEUDO-RESPONSE REGULATORs[J]. Plant Cell Physiol, 2016, 57(5): 1085-1097.
doi: 10.1093/pcp/pcw057 pmid: 27012548 |
| [25] | 向芬, 黄帅, 赵小英, 等. 拟南芥prr5突变体对ABA 的响应[J]. 激光生物学报, 2013, 22(6): 546-550. |
| Xiang F, Huang S, Zhao XY, et al. The response of the prr5 mutant to ABA in Arabidopsis thaliana[J]. Acta Laser Biology Sinica, 2013, 22(6): 546-550. | |
| [26] |
Nakamichi N, Kita M, Niinuma K, et al. Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway[J]. Plant Cell Physiol, 2007, 48(6): 822-832.
doi: 10.1093/pcp/pcm056 pmid: 17504813 |
| [27] |
Hayama R, Sarid-Krebs L, Richter R, et al. PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length[J]. EMBO J, 2017, 36(7): 904-918.
doi: 10.15252/embj.201693907 pmid: 28270524 |
| [28] |
贾小平, 李剑峰, 张博, 等. 谷子SiPRR37基因对光温、非生物胁迫的响应特点及其有利等位变异鉴定[J]. 作物学报, 2021, 47(4): 638-649.
doi: 10.3724/SP.J.1006.2021.04139 |
| Jia XP, Li JF, Zhang B, et al. Responsive features of SiPRR37 to photoperiod and temperature, abiotic stress and identification of its favourable allelic variations in foxtail millet(Setaria italica L.)[J]. Acta Agron Sin, 2021, 47(4): 638-649. | |
| [29] | 王玉岚. 生物钟基因OsPRR37耐旱功能的研究及其编辑在水稻育种中的应用[D]. 武汉: 华中农业大学, 2022. |
| Wang YL. Study on drought resistance function of biological clock gene OsPRR37 and application of gene editing in rice breeding[D]. Wuhan: Huazhong Agricultural University, 2022. | |
| [30] |
Gao H, Jin MN, Zheng XM, et al. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice[J]. Proc Natl Acad Sci USA, 2014, 111(46): 16337-16342.
doi: 10.1073/pnas.1418204111 pmid: 25378698 |
| [31] | Li CC, Ma J, Wang GP, et al. Exploring the SiCCT gene family and its role in heading date in foxtail millet[J]. Front Plant Sci, 2022, 13: 863298. |
| [32] |
Campoli C, Shtaya M, Davis SJ, et al. Expression conservation within the circadian clock of a monocot: natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs[J]. BMC Plant Biol, 2012, 12: 97.
doi: 10.1186/1471-2229-12-97 pmid: 22720803 |
| [33] |
Yamamoto Y, Sato E, Shimizu T, et al. Comparative genetic studies on the APRR5 and APRR7 genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis[J]. Plant Cell Physiol, 2003, 44(11): 1119-1130.
pmid: 14634148 |
| [34] | Ito S, Kawamura H, Niwa Y, et al. A genetic study of the Arabidopsis circadian clock with reference to the TIMING OF CAB EXPRESSION 1(TOC1)gene[J]. Plant Cell Physiol, 2009, 50(2): 290-303. |
| [35] | Pin PA, Nilsson O. The multifaceted roles of FLOWERING LOCUS T in plant development[J]. Plant Cell Environ, 2012, 35(10): 1742-1755. |
| [36] |
Wang LW, Sun S, Wu TT, et al. Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean[J]. Plant Biotechnol J, 2020, 18(9): 1869-1881.
doi: 10.1111/pbi.13346 pmid: 31981443 |
| [1] | 李雨晴, 吴楠, 罗建让. 卵叶牡丹花色苷合成相关基因bHLH的克隆与功能分析[J]. 生物技术通报, 2024, 40(8): 174-185. |
| [2] | 崔原瑗, 王昭懿, 白双宇, 任毓昭, 豆飞飞, 刘彩霞, 刘凤楼, 王掌军, 李清峰. 大麦非特异性磷脂酶C基因家族全基因组鉴定及苗期胁迫表达分析[J]. 生物技术通报, 2024, 40(8): 74-82. |
| [3] | 杨巍, 赵丽芬, 唐兵, 周麟笔, 杨娟, 莫传园, 张宝会, 李飞, 阮松林, 邓英. 芥菜SRO基因家族全基因组鉴定与表达分析[J]. 生物技术通报, 2024, 40(8): 129-141. |
| [4] | 周冉, 王兴平, 李彦霞, 罗仍卓么. 金黄色葡萄球菌型乳房炎奶牛乳腺组织的lncRNA差异表达分析[J]. 生物技术通报, 2024, 40(8): 320-328. |
| [5] | 张明亚, 庞胜群, 刘玉东, 苏永峰, 牛博文, 韩琼琼. 番茄FAD基因家族的鉴定与表达分析[J]. 生物技术通报, 2024, 40(7): 150-162. |
| [6] | 臧文蕊, 马明, 砗根, 哈斯阿古拉. 甜瓜BZR转录因子家族基因的全基因组鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(7): 163-171. |
| [7] | 杨嘉泓, 李婧怡, 吴佳昊, 黄幼梅, 刘艳芬, 秦源, 蔡汉阳. 生长素信号途径参与调控拟南芥雌配子体发育研究进展[J]. 生物技术通报, 2024, 40(7): 19-27. |
| [8] | 杜仲阳, 杨泽, 梁梦静, 刘义珍, 崔红利, 史达明, 薛金爱, 孙岩, 张春辉, 季春丽, 李润植. 纳米硒(SeNPs)缓解烟草幼苗铅胁迫和促生效应[J]. 生物技术通报, 2024, 40(7): 183-196. |
| [9] | 阿丽亚·外力, 陈永坤, 克拉热木·克里木江, 王宝庆, 陈凌娜. 核桃SPL基因家族的系统进化和表达分析[J]. 生物技术通报, 2024, 40(6): 180-189. |
| [10] | 常雪瑞, 王田田, 王静. 辣椒E2基因家族的鉴定及分析[J]. 生物技术通报, 2024, 40(6): 238-250. |
| [11] | 刘蓉, 田闵玉, 李光泽, 谭成方, 阮颖, 刘春林. 甘蓝型油菜REVEILLE家族鉴定及诱导表达分析[J]. 生物技术通报, 2024, 40(6): 161-171. |
| [12] | 王玉书, 赵琳琳, 赵爽, 胡琦, 白慧霞, 王欢, 曹业萍, 范震宇. 大白菜BrCYP83B1基因的克隆及表达分析[J]. 生物技术通报, 2024, 40(6): 152-160. |
| [13] | 李博静, 郑腊梅, 吴乌云, 高飞, 周宜君. 西蒙得木HSP20基因家族的进化、表达和功能分析[J]. 生物技术通报, 2024, 40(6): 190-202. |
| [14] | 郝思怡, 张君珂, 王斌, 曲朋燕, 李瑞得, 程春振. 香蕉ELF3的克隆与表达分析[J]. 生物技术通报, 2024, 40(5): 131-140. |
| [15] | 李嘉欣, 李鸿燕, 刘丽娥, 张恬, 周武. 沙棘NRAMP基因家族鉴定及铅胁迫下表达分析[J]. 生物技术通报, 2024, 40(5): 191-202. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||