








生物技术通报 ›› 2025, Vol. 41 ›› Issue (2): 150-162.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0689
• 研究报告 • 上一篇
许圆梦1,2,3( ), 毛娇1,2,3, 王梦瑶1,2,3, 王数1,2,3, 任江陵1,2,3, 刘宇涵1,2,3, 刘思辰1,2,3, 乔治军1,2,3, 王瑞云1(
), 毛娇1,2,3, 王梦瑶1,2,3, 王数1,2,3, 任江陵1,2,3, 刘宇涵1,2,3, 刘思辰1,2,3, 乔治军1,2,3, 王瑞云1( ), 曹晓宁1,2,3(
), 曹晓宁1,2,3( )
)
收稿日期:2024-07-18
出版日期:2025-02-26
发布日期:2025-02-28
通讯作者:
王瑞云,女,博士,教授,研究方向 :糜子分子育种;E-mail: wry925@126.com作者简介:许圆梦,女,硕士研究生,研究方向 :糜子分子育种;E-mail: xuyuanmeng1229@163.com
基金资助:
XU Yuan-meng1,2,3( ), MAO Jiao1,2,3, WANG Meng-yao1,2,3, WANG Shu1,2,3, REN Jiang-ling1,2,3, LIU Yu-han1,2,3, LIU Si-chen1,2,3, QIAO Zhi-jun1,2,3, WANG Rui-yun1(
), MAO Jiao1,2,3, WANG Meng-yao1,2,3, WANG Shu1,2,3, REN Jiang-ling1,2,3, LIU Yu-han1,2,3, LIU Si-chen1,2,3, QIAO Zhi-jun1,2,3, WANG Rui-yun1( ), CAO Xiao-ning1,2,3(
), CAO Xiao-ning1,2,3( )
)
Received:2024-07-18
Published:2025-02-26
Online:2025-02-28
摘要:
目的 穗型是影响作物产量和机械化收割的重要因素,DEP1(Dense Erect Panicles 1)和EP3(Erect Panicle 3)基因是控制穗型形成的关键基因。旨在探究糜子(Panicum miliaceum L.)PmDEP1和PmEP3基因的结构与表达特征。 方法 以散穗型糜子笤帚糜子(ZM)、密穗型糜子M278和侧穗型糜子M350为材料,克隆获得PmDEP1和PmEP3,并对其开展生物信息学分析。采用RT-qPCR检测不同穗型糜子PmDEP1和PmEP3的表达模式。 结果 序列结果分析显示,PmDEP1 cDNA全长为1 044 bp,编码347个氨基酸,蛋白结构域预测为PAT1,二级结构以无规则卷曲为主,三级结构与水稻DEP1蛋白高度相似,预测定位于细胞核,与柳枝稷具有较高的同源性,拥有28个磷酸化位点,不具有跨膜结构和信号肽。PmEP3 cDNA全长1 215 bp,编码358个氨基酸,编码的蛋白属于F-box家族,二级结构以无规则卷曲和延伸链为主,三级结构与水稻Os02g0260200蛋白高度相似,预测定位于细胞质,与柳枝稷具有较高的同源性,不含信号肽,拥有32个磷酸化位点,具有少量的跨膜结构。RT-PCR结果显示,PmDEP1和PmEP3在不同时期的不同部位中表达,PmDEP1在拔节期的根中表达量最高,在抽穗期的笤帚糜子和M278中的叶部表达量最高,在M350中的茎部表达量最高。PmEP3在拔节期的叶中表达量最高,在笤帚糜子和M278抽穗期的叶部表达量最高,在M350的穗部表达量最高。 结论 PmDEP1和PmEP3为穗型基因,可能参与调控糜子穗型形成。
许圆梦, 毛娇, 王梦瑶, 王数, 任江陵, 刘宇涵, 刘思辰, 乔治军, 王瑞云, 曹晓宁. 糜子PmDEP1和PmEP3基因的克隆与表达特征分析[J]. 生物技术通报, 2025, 41(2): 150-162.
XU Yuan-meng, MAO Jiao, WANG Meng-yao, WANG Shu, REN Jiang-ling, LIU Yu-han, LIU Si-chen, QIAO Zhi-jun, WANG Rui-yun, CAO Xiao-ning. Cloning and Expression Characteristics Analysis of Millet Genes PmDEP1 and PmEP3[J]. Biotechnology Bulletin, 2025, 41(2): 150-162.
| 引物 Primer | 引物序列 Primer sequence (5′-3′) | 功能 Primer function |
|---|---|---|
| actin | TCTGTTTTTCTCTATGTGCAGGA | 基因表达分析 Analysis of gene expression |
| PmDEP1-Q | TGCTGCTTCCATTGTTCTTGCTG | |
| PmEP3-Q | GAGCCTCCGCAAGTGGTATGG | |
| PmDEP1 | AGTGAATTCGAGCTCGGTACATGGGGGAGGAGGTGGCG | 基因克隆 Gene cloning |
| PmEP3 | AGTGAATTCGAGCTCGGTACATGGGGTCGGAAGAGTGGGAG |
表1 PmDEP1和PmEP3的引物序列
Table 1 Primer sequences for PmDEP1 and PmEP3
| 引物 Primer | 引物序列 Primer sequence (5′-3′) | 功能 Primer function |
|---|---|---|
| actin | TCTGTTTTTCTCTATGTGCAGGA | 基因表达分析 Analysis of gene expression |
| PmDEP1-Q | TGCTGCTTCCATTGTTCTTGCTG | |
| PmEP3-Q | GAGCCTCCGCAAGTGGTATGG | |
| PmDEP1 | AGTGAATTCGAGCTCGGTACATGGGGGAGGAGGTGGCG | 基因克隆 Gene cloning |
| PmEP3 | AGTGAATTCGAGCTCGGTACATGGGGTCGGAAGAGTGGGAG |
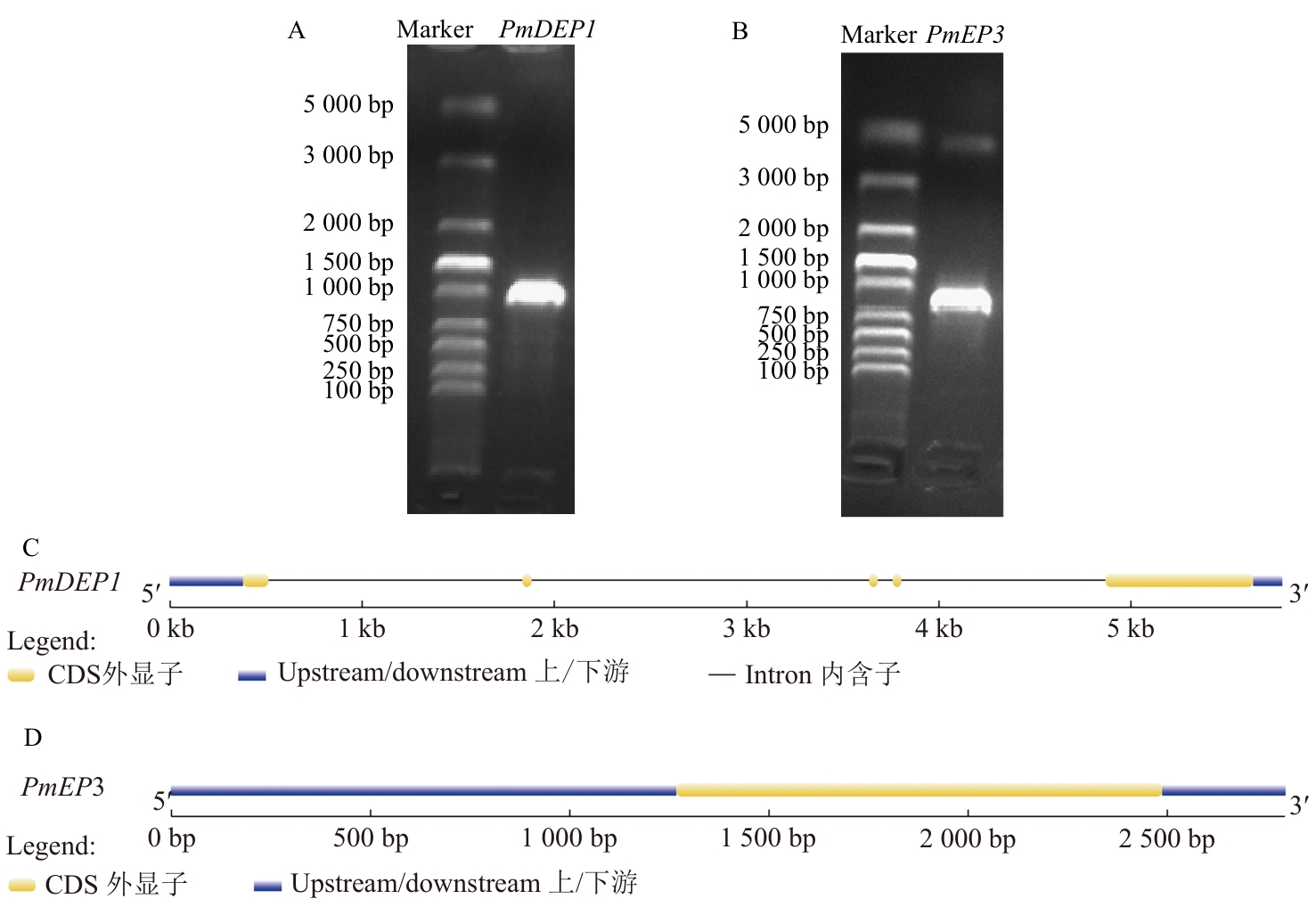
图1 PmDEP1和PmEP3基因的克隆和结构分析A:PmDEP1的凝胶电泳图;B:PmEP3的凝胶电泳图;C:PmDEP1基因结构;D:PmEP3基因结构
Fig. 1 Cloning and structural analysis of PmDEP1 and PmEP3 genesA: Gel electrophoresis map of PmDEP1 gene. B: Gel electrophoresis map of PmEP3 gene. C: PmDEP1 gene structure. D: PmEP3 gene structure

图2 PmDEP1和PmEP3的核苷酸及氨基酸序列和基因结构域A:PmDEP1的核苷酸及氨基酸序列;B:PmEP3的核苷酸及氨基酸序列;C:PmDEP1基因结构域;D:PmEP3基因结构域
Fig. 2 Nucleotide and amino acid sequences and gene domains of PmDEP1 and PmEP3A: Nucleotide and amino acid sequence of PmDEP1. B: Nucleotide and amino acid sequence of PmEP3. C: PmDEP1 gene domain. D: PmEP3 gene domain
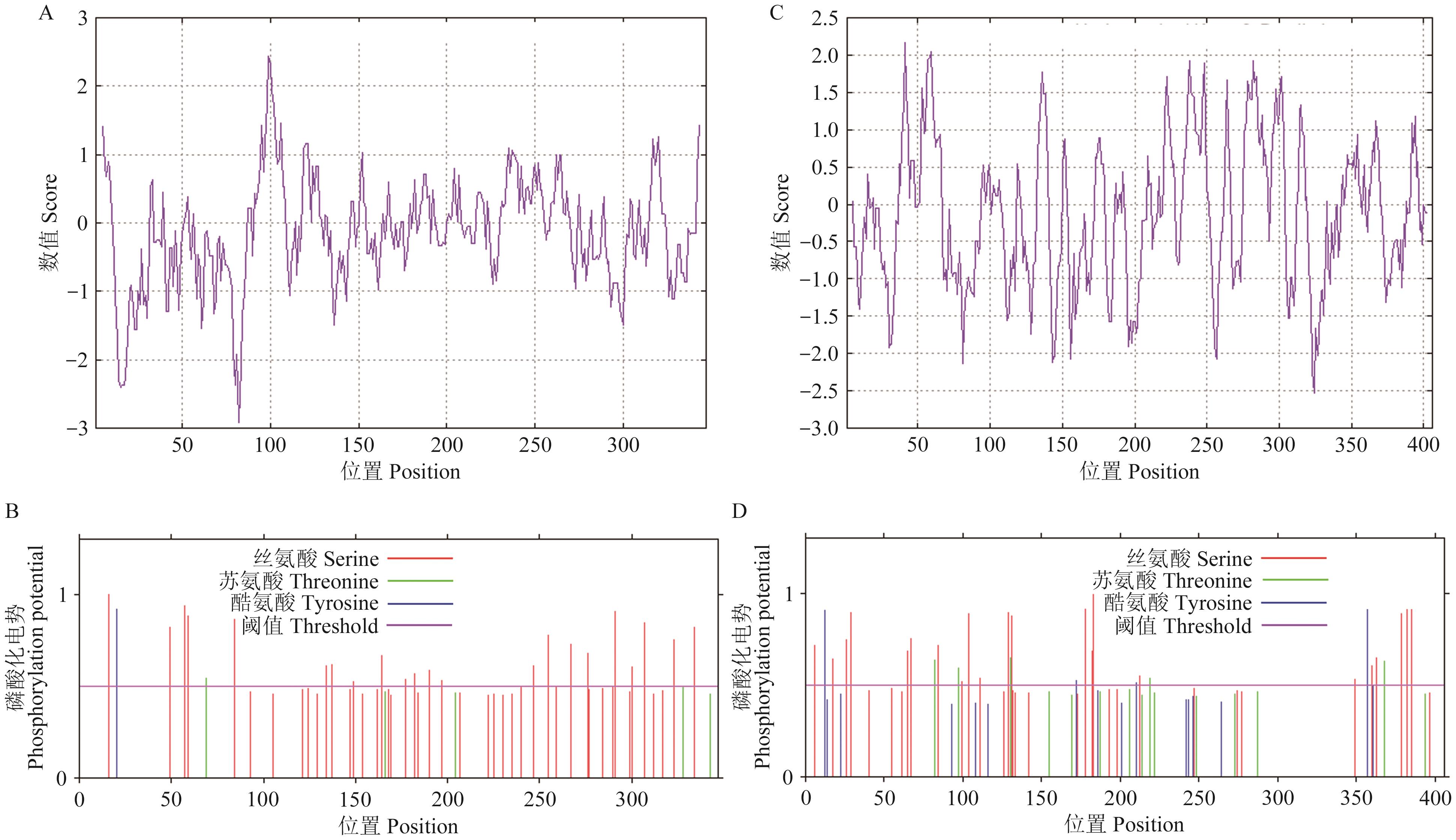
图4 PmDEP1和PmEP3蛋白的疏水性/亲水性和磷酸化预测A:PmDEP1蛋白的疏水性/亲水性预测;B:PmDEP1蛋白的磷酸化预测;C:PmEP3蛋白的疏水性/亲水性预测;D:PmEP3蛋白的磷酸化预测
Fig. 4 Prediction of hydrophobicity/hydrophilicity and phosphorylation of PmDEP1 and PmEP3 proteinsA: Hydrophobic/hydrophilic prediction of PmDEP1 protein. B: Phosphorylation prediction of PmDEP1 protein. C: Hydrophobic/hydrophilic prediction of PmEP3 protein. D: Phosphorylation prediction of PmEP3 protein
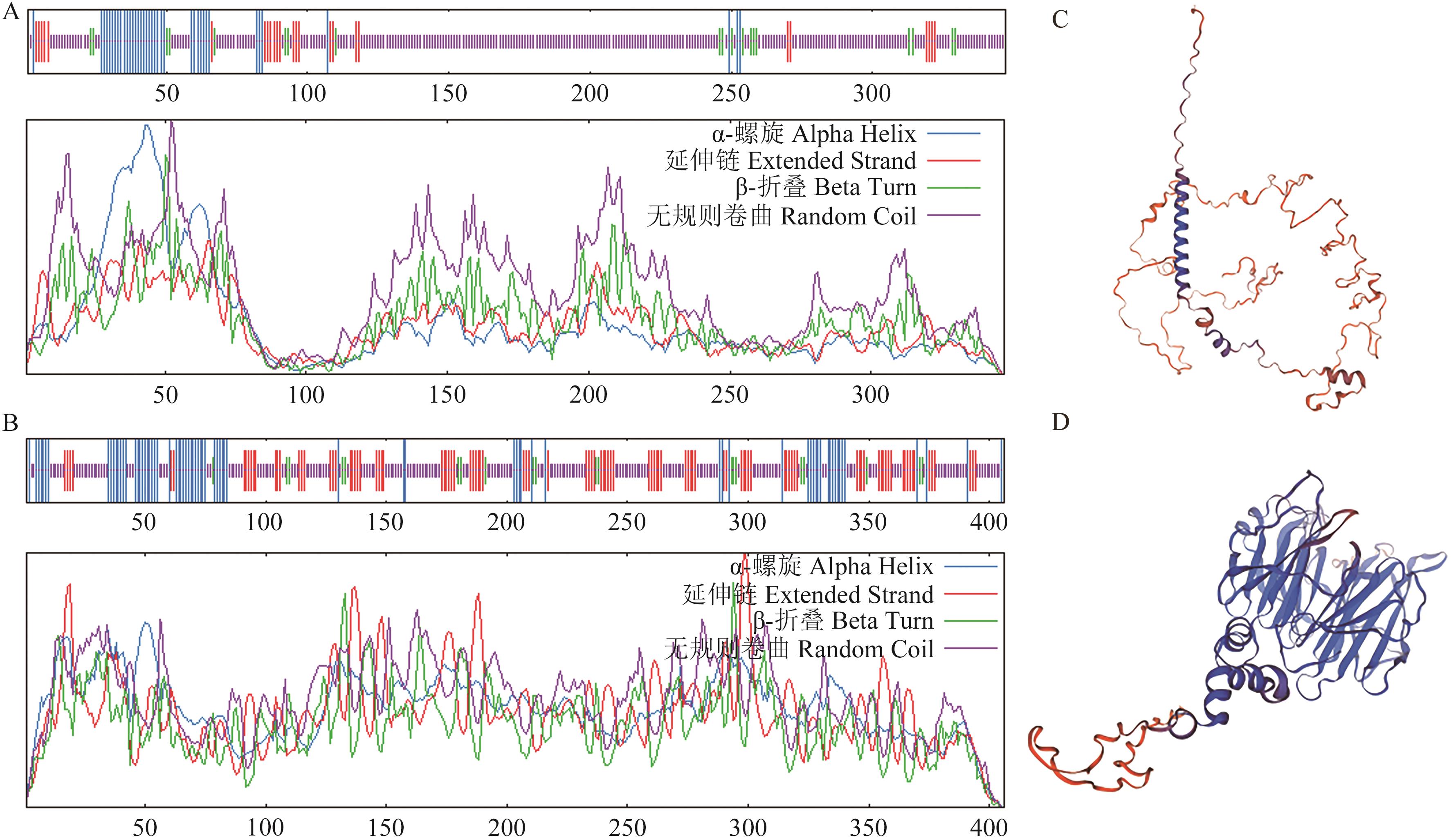
图7 PmDEP1蛋白和PmEP3蛋白二级结构和三级结构预测A:PmDEP1蛋白的二级结构;B:PmEP3蛋白的二级结构;C:PmDEP1蛋白的三级结构;D:PmEP3蛋白的三级结构
Fig. 7 Prediction of secondary and tertiary structures of PmDEP1 and PmEP3 proteinsA: Secondary structure of PmDEP1 protein. B: Secondary structure of PmEP3 protein. C: Tertiary structure of PmDEP1 protein. D: Tertiary structure of PmEP3 protein

图8 PmDEP1蛋白和PmEP3蛋白信号肽和跨膜结构域预测A:PmDEP1蛋白信号肽预测;B:PmEP3蛋白信号肽预测;C:PmDEP1蛋白跨膜结构域预测;D:PmEP3跨膜结构域预测
Fig. 8 Signal peptide and transmembrane domain prediction of PmDEP1 protein and PmEP3 proteinA: Prediction of PmDEP1 protein signal peptide. B: Prediction of PmEP3 protein signal peptide. C: Prediction of PmDEP1 protein transmembrane domain. D: Prediction of PmEP3 transmembrane domain
亚细胞位置 Subcellular location | 可能性 Possibility/% | ||
|---|---|---|---|
| PmDEP1 | PmEP3 | ||
| 细胞核 | 78.3 | 26.1 | |
| 细胞质 | 8.7 | 47.8 | |
| 质膜 | 4.3 | - | |
| 过氧化物酶体 | 4.3 | - | |
| 线粒体 | 4.3 | 26.1 | |
表2 PmDEP1和PmEP3蛋白的亚细胞定位预测结果
Table 2 Prediction results of subcellular localizations for PmDEP1 and PmEP3 proteins
亚细胞位置 Subcellular location | 可能性 Possibility/% | ||
|---|---|---|---|
| PmDEP1 | PmEP3 | ||
| 细胞核 | 78.3 | 26.1 | |
| 细胞质 | 8.7 | 47.8 | |
| 质膜 | 4.3 | - | |
| 过氧化物酶体 | 4.3 | - | |
| 线粒体 | 4.3 | 26.1 | |
基因名称 Gene name | 作用元件 Functional component | 核心序列 Core sequence | 数量 Number | 功能 Function |
|---|---|---|---|---|
| PmDEP1 | CGTCA-motif | CGTCA | 1 | MeJA作用调控元件 |
| TGACG-motif | TGACG | 1 | MeJA作用调控元件 | |
| G-box | CACGAC | 4 | 光响应应答元件 | |
| chs-CMA2a | TCACTTGA | 1 | 光响应应答元件 | |
| TCA-element | CCATCTTTTT | 1 | 水杨酸作用元件 | |
| P-box | CCTTTTG | 1 | 赤霉素反应元件 | |
| ARE | AAACCA | 1 | 厌氧诱导调控元件 | |
| GC-motif | CCCCCG | 2 | 参与缺氧特异性诱导元件 | |
| MBS | CAACTG | 1 | 干旱诱导元件 | |
| CCGTCC-box | CCGTCC | 1 | 与分生组织特异性激活调节元件 | |
| STRE | AGGGG | 4 | 应激反应元件 | |
| PmEP3 | CGTCA-motif | CGTCA | 1 | MeJA作用调控元件 |
| TGACG-motif | TGACG | 1 | MeJA作用调控元件 | |
| ATCT-motif | AATCTAATCC | 1 | 光响应应答元件 | |
| TCT-motif | TCTTAC | 2 | 光响应应答元件 | |
| MRE | AACCTAA | 1 | MYB参与光反应结合位点 | |
| GARE-motif | TCTGTTG | 1 | 赤霉素反应元件 | |
| TC-rich repeats | ATTCTCTAAC | 1 | 赤霉素反应元件 | |
| AuxRR-core | GGTCCAT | 1 | 生长素反应调控元件 | |
| AAGAA-motif | GAAAGAA | 1 | 脱落酸反应性相关性元件 | |
| ARE | AAACCA | 2 | 厌氧诱导调控元件 | |
| MBS | CAACTG | 1 | 干旱诱导元件 | |
| CAT-box | GCCACT | 1 | 分生组织表达调控元件 | |
| GCN4_motif | TGAGTCA | 1 | 参与胚乳表达的顺式调控元件 |
表3 PmDEP1和PmEP3启动子顺式作用元件分析
Table 3 Analysis of cis-acting elements of PmDEP1 and PmEP3 promoters
基因名称 Gene name | 作用元件 Functional component | 核心序列 Core sequence | 数量 Number | 功能 Function |
|---|---|---|---|---|
| PmDEP1 | CGTCA-motif | CGTCA | 1 | MeJA作用调控元件 |
| TGACG-motif | TGACG | 1 | MeJA作用调控元件 | |
| G-box | CACGAC | 4 | 光响应应答元件 | |
| chs-CMA2a | TCACTTGA | 1 | 光响应应答元件 | |
| TCA-element | CCATCTTTTT | 1 | 水杨酸作用元件 | |
| P-box | CCTTTTG | 1 | 赤霉素反应元件 | |
| ARE | AAACCA | 1 | 厌氧诱导调控元件 | |
| GC-motif | CCCCCG | 2 | 参与缺氧特异性诱导元件 | |
| MBS | CAACTG | 1 | 干旱诱导元件 | |
| CCGTCC-box | CCGTCC | 1 | 与分生组织特异性激活调节元件 | |
| STRE | AGGGG | 4 | 应激反应元件 | |
| PmEP3 | CGTCA-motif | CGTCA | 1 | MeJA作用调控元件 |
| TGACG-motif | TGACG | 1 | MeJA作用调控元件 | |
| ATCT-motif | AATCTAATCC | 1 | 光响应应答元件 | |
| TCT-motif | TCTTAC | 2 | 光响应应答元件 | |
| MRE | AACCTAA | 1 | MYB参与光反应结合位点 | |
| GARE-motif | TCTGTTG | 1 | 赤霉素反应元件 | |
| TC-rich repeats | ATTCTCTAAC | 1 | 赤霉素反应元件 | |
| AuxRR-core | GGTCCAT | 1 | 生长素反应调控元件 | |
| AAGAA-motif | GAAAGAA | 1 | 脱落酸反应性相关性元件 | |
| ARE | AAACCA | 2 | 厌氧诱导调控元件 | |
| MBS | CAACTG | 1 | 干旱诱导元件 | |
| CAT-box | GCCACT | 1 | 分生组织表达调控元件 | |
| GCN4_motif | TGAGTCA | 1 | 参与胚乳表达的顺式调控元件 |
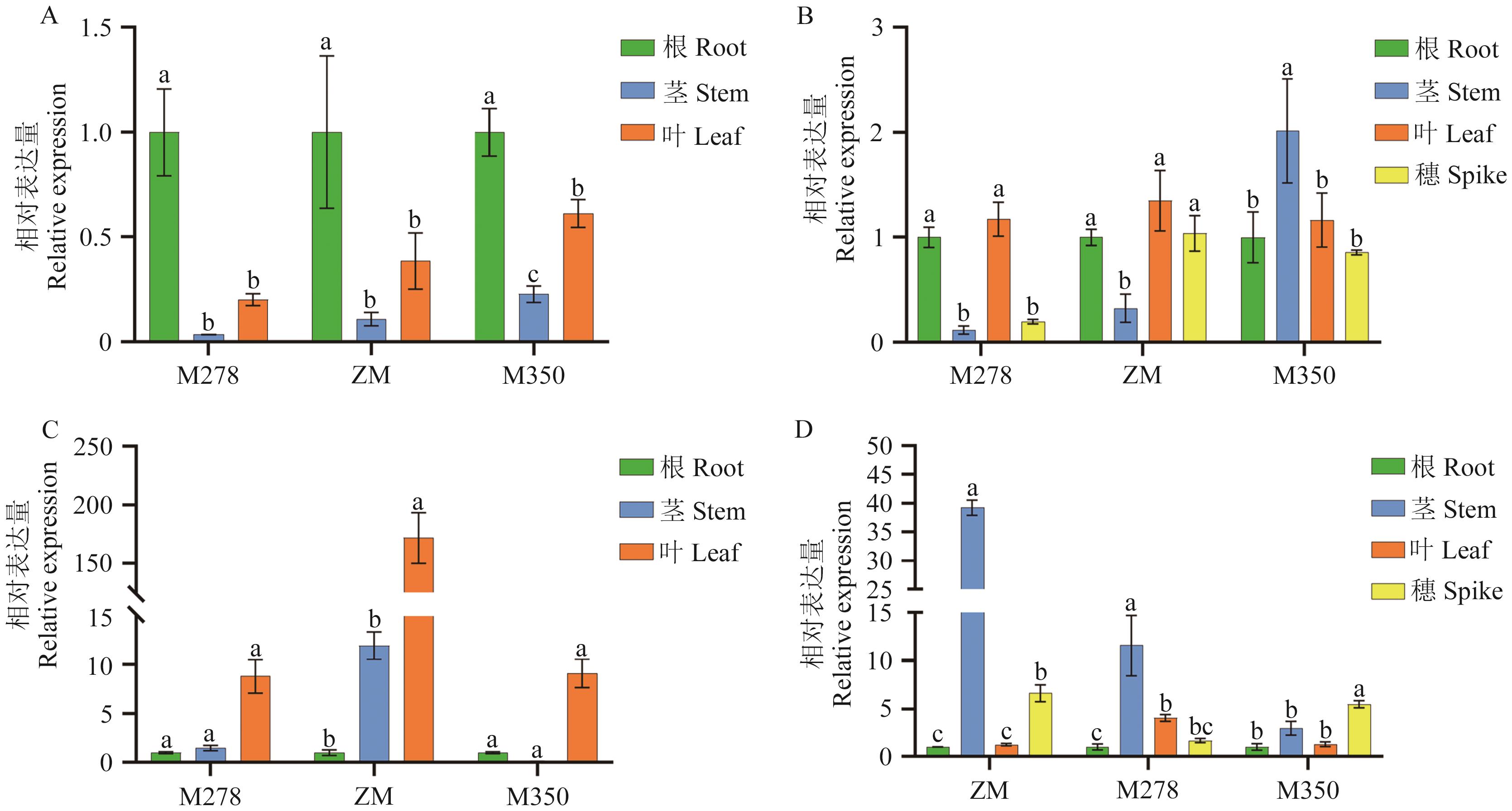
图10 糜子PmDEP1和PmEP3在ZM、M350和M278的拔节期和抽穗期的不同部位表达量A:PmDEP1在拔节期各个部位表达情况;B:PmDEP1在抽穗期各个部位表达情况;C:PmEP3在拔节期各个部位表达情况;D:PmEP3在抽穗期各个部位表达情况;不同小写字母表示表达量在P<0.05水平上差异显著
Fig. 10 Expressions of PmDEP1 and PmEP3 genes in different parts of ZM, M350 and M278 at jointing and heading stagesA: Expressions of PmDEP1 in different parts at jointing stage. B: Expressions of PmDEP1 in different parts at heading stage. C: Expressions of PmEP3 in different parts at jointing stage. D: Expressions of PmEP3 in various parts at heading stage. Different lowercase letters indicate that the expression was significantly different at the P<0.05 level
| 1 | Boukail S, Macharia M, Miculan M, et al. Genome wide association study of agronomic and seed traits in a world collection of proso millet (Panicum miliaceum L.) [J]. BMC Plant Biol, 2021, 21(1): 330. |
| 2 | Yuan YH, Liu CJ, Gao YB, et al. Proso millet (Panicum miliaceum L.): a potential crop to meet demand scenario for sustainable saline agriculture [J]. J Environ Manage, 2021, 296: 113216. |
| 3 | 王纶, 王星玉, 王海岗, 等. 山西重要黍稷种质资源品质性状的初步鉴定与评价 [J]. 植物遗传资源学报, 2017, 18(1): 61-69. |
| Wang L, Wang XY, Wang HG, et al. Preliminary appraisal of important proso mille germplasm resources quality traits in Shanxi Province [J]. J Plant Genet Resour, 2017, 18(1): 61-69. | |
| 4 | 刁现民. 禾谷类杂粮作物耐逆和栽培技术研究新进展[J]. 中国农业科学, 2019, 52(22): 3943-3949. |
| Diao XM. New research progress on stress tolerance and cultivation techniques of cereal crops[J]. Sci Agric Sin, 2019, 52(22): 3943-3949. | |
| 5 | 曹晓宁, 王君杰, 刘思辰, 等. 密穗型糜子资源的筛选评价 [J] . 现代农业科技, 2016, 18:37, 41. |
| Cao XN, Wang JJ, Liu SC, et al. Screening and evaluation of dense-spike broomcorn millet resources [J] . Mod Agric Sci Technol, 2016, 18:37, 41. | |
| 6 | 王纶, 王星玉, 乔治军, 等. 黍稷种质穗型与主要农艺性状的关系 [J]. 山西农业科学, 2013, 41(8): 789-792, 796. |
| Wang L, Wang XY, Qiao ZJ, et al. Study on broomcorn millet germplasm panicle type and its main agronomic traits [J]. J Shanxi Agric Sci, 2013, 41(8): 789-792, 796. | |
| 7 | 唐勇. 酿酒高粱SbGA2o3基因的TILLING筛选及抗旱性分析 [D]. 贵阳: 贵州大学, 2022. |
| Tang Y. TILLING screening and drought resistance analysis of SbGA2o3 gene in brewing sorghum [D]. Guiyang: Guizhou University, 2022. | |
| 8 | Fei C, Yu JH, Xu ZJ, et al. Erect panicle architecture contributes to increased rice production through the improvement of canopy structure [J]. Mol Breed, 2019, 39(9): 128. |
| 9 | Hirooka Y, Homma K, Shiraiwa T, et al. Yield and growth characteristics of erect panicle type rice (Oryza sativaL.) cultivar, Shennong265 under various crop management practices in Western Japan [J]. Plant Prod Sci, 2018, 21(1): 1-7. |
| 10 | 张喜娟, 李红娇, 李伟娟, 等. 北方直立穗型粳稻抗倒性的研究 [J]. 中国农业科学, 2009, 42(7): 2305-2313. |
| Zhang XJ, Li HJ, Li WJ, et al. The lodging resistance of erect panicle Japonica rice in northern China [J]. Sci Agric Sin, 2009, 42(7): 2305-2313. | |
| 11 | Yang YH, Shen ZY, Xu CD, et al. Genetic improvement of panicle-erectness Japonica rice toward both yield and eating and cooking quality [J]. Mol Breed, 2020, 40(5): 51. |
| 12 | Oba GC, Goneli ALD, Masetto TE, et al. Artificial drying of safflower seeds at different air temperatures: effect on the physiological potential of freshly harvested and stored seeds [J]. J Seed Sci, 2019, 41(4): 397-406. |
| 13 | Kong FN, Wang JY, Zou JC, et al. Molecular tagging and mapping of the erect panicle gene in rice [J]. Mol Breed, 2007, 19(4): 297-304. |
| 14 | Li SJ, Liu WX, Zhang XQ, et al. Roles of the Arabidopsis G protein γ subunit AGG3 and its rice homologs GS3 and DEP1 in seed and organ size control [J]. Plant Signal Behav, 2012, 7(10): 1357-1359. |
| 15 | Huang XZ, Qian Q, Liu ZB, et al. Natural variation at the DEP1 locus enhances grain yield in rice [J]. Nat Genet, 2009, 41(4): 494-497. |
| 16 | 黄勇, 胡勇, 傅向东, 等. 水稻产量性状的功能基因及其应用 [J]. 生命科学, 2016, 28(10): 1147-1155. |
| Huang Y, Hu Y, Fu XD, et al. Functional genes for grain yield related traits and their application in rice breeding [J]. Chin Bull Life Sci, 2016, 28(10): 1147-1155. | |
| 17 | Ashikari M, Sakakibara H, Lin SY, et al. Cytokinin oxidase regulates rice grain production [J]. Science, 2005, 309(5735): 741-745. |
| 18 | Wendt T, Holme I, Dockter C, et al. HvDep1 is a positive regulator of culm elongation and grain size in barley and impacts yield in an environment-dependent manner [J]. PLoS One, 2016, 11(12): e0168924. |
| 19 | Piao RH, Jiang WZ, Ham TH, et al. Map-based cloning of the ERECT PANICLE 3 gene in rice [J]. Theor Appl Genet, 2009, 119(8): 1497-1506. |
| 20 | Yu HY, Murchie EH, González-Carranza ZH, et al. Decreased photosynthesis in the erect panicle 3 (ep3) mutant of rice is associated with reduced stomatal conductance and attenuated guard cell development [J]. J Exp Bot, 2015, 66(5): 1543-1552. |
| 21 | Li M, Tang D, Wang KJ, et al. Mutations in the F-box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice [J]. Plant Biotechnol J, 2011, 9(9): 1002-1013. |
| 22 | Borna RS, Murchie EH, Pyke KA, et al. The rice EP3 and OsFBK1 E3 ligases alter plant architecture and flower development, and affect transcript accumulation of microRNA pathway genes and their targets [J]. Plant Biotechnol J, 2022, 20(2): 297-309. |
| 23 | 靳丰璐. 基于转录组测序对糜子直立穗突变体的分子机制研究 [D]. 太谷: 山西农业大学, 2022. |
| Jin FL. The study on the molecular mechanism of the erect panicle mutant in broomcorn millet based on the transcriptome sequencing analysis [D]. Taigu: Shanxi Agricultural University, 2022. | |
| 24 | Li F, Liu WB, Tang JY, et al. Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation [J]. Cell Res, 2010, 20(7): 838-849. |
| 25 | Abe Y, Mieda K, Ando T, et al. The SMALL AND ROUND SEED1 (SRS1/DEP2) gene is involved in the regulation of seed size in rice [J]. Genes Genet Syst, 2010, 85(5): 327-339. |
| 26 | Qiao YL, Piao RH, Shi JX, et al. Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.) [J]. Theor Appl Genet, 2011, 122(7): 1439-1449. |
| 27 | Zhu KM, Tang D, Yan CJ, et al. Erect panicle2 encodes a novel protein that regulates panicle erectness in indica rice [J]. Genetics, 2010, 184(2): 343-350. |
| 28 | Zhou Y, Zhu JY, Li ZY, et al. Deletion in a quantitative trait gene qPE9-1 associated with panicle erectness improves plant architecture during rice domestication [J]. Genetics, 2009, 183(1): 315-324. |
| 29 | 刘亚男, 夏先春, 何中虎. 普通小麦TaDep1基因克隆与特异性标记开发 [J]. 作物学报, 2013, 39(4): 589-598. |
| Liu YN, Xia XC, He ZH. Characterization of dense and erect panicle 1 gene (TaDep1) located on common wheat group 5 chromosomes and development of allele-specific markers [J]. Acta Agron Sin, 2013, 39(4): 589-598. | |
| 30 | 黄幸, 丁峰, 潘介春, 等. 龙眼DlWRKY57基因克隆与拟南芥遗传转化 [J]. 分子植物育种, 2021, 19(1): 149-157. |
| Huang X, Ding F, Pan JC, et al. Cloning and Arabidopsis genetic transformation of DlWRKY57 gene in Dimocarpus longan [J]. Mol Plant Breed, 2021, 19(1): 149-157. | |
| 31 | Liu J, Wang XY, Chen YL, et al. Identification, evolution and expression analysis of WRKY gene family in Eucommia ulmoides [J]. Genomics, 2021, 113(5): 3294-3309. |
| 32 | Wang JY, Nakazaki T, Chen SQ, et al. Identification and characterization of the erect-pose panicle gene EP conferring high grain yield in rice (Oryza sativa L.) [J]. Theor Appl Genet, 2009, 119(1): 85-91. |
| 33 | 黄先忠. 水稻直立密穗控制基因dep1的克隆和功能研究 [D]. 北京: 中国科学院研究生院, 2009. |
| Huang XZ. Cloning and functional study of the control gene dep1 for erect dense panicle in rice [D]. Beijing: Graduate University of Chinese Academy of Sciences, 2009. | |
| 34 | 吴怀通. 棉花纤维起始发育基因Li3 的图位克隆和功能验证 [D]. 南京: 南京农业大学, 2017. |
| Wu HT. Mapping cloning and functional verification of cotton fiber initiation development gene Li3 [D]. Nanjing: Nanjing Agricultural University, 2017. |
| [1] | 马天意, 许家佳, 路文婧, 吴艳, 沙伟, 张梅娟, 彭疑芳. ‘金小童’大白菜BrcGASA3基因在盐碱胁迫下的表达分析及抗性鉴定[J]. 生物技术通报, 2025, 41(2): 127-138. |
| [2] | 李禹欣, 李苗, 杜晓芬, 韩康妮, 连世超, 王军. 谷子SiSAP基因家族的鉴定与表达分析[J]. 生物技术通报, 2025, 41(1): 143-156. |
| [3] | 孔青洋, 张晓龙, 李娜, 张晨洁, 张雪云, 于超, 张启翔, 罗乐. 单叶蔷薇GRAS转录因子家族鉴定及表达分析[J]. 生物技术通报, 2025, 41(1): 210-220. |
| [4] | 宋兵芳, 柳宁, 程新艳, 徐晓斌, 田文茂, 高悦, 毕阳, 王毅. 马铃薯G6PDH基因家族鉴定及其在损伤块茎的表达分析[J]. 生物技术通报, 2024, 40(9): 104-112. |
| [5] | 吴慧琴, 王延宏, 刘涵, 司政, 刘雪晴, 王静, 阳宜, 成妍. 辣椒UGT基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(9): 198-211. |
| [6] | 满全财, 孟姿诺, 李伟, 蔡心汝, 苏润东, 付长青, 高顺娟, 崔江慧. 马铃薯AQP基因家族鉴定及表达分析[J]. 生物技术通报, 2024, 40(9): 51-63. |
| [7] | 乔岩, 杨芳, 任盼荣, 祁伟亮, 安沛沛, 李茜, 李丹, 肖俊飞. 马铃薯野生种烯酰水合酶超家族基因ScDHNS的克隆与功能分析[J]. 生物技术通报, 2024, 40(9): 92-103. |
| [8] | 申鹏, 高雅彬, 丁红. 马铃薯SAT基因家族的鉴定和表达分析[J]. 生物技术通报, 2024, 40(9): 64-73. |
| [9] | 李亦君, 杨小贝, 夏琳, 罗朝鹏, 徐馨, 杨军, 宁黔冀, 武明珠. 烟草NtPRR37基因克隆及功能分析[J]. 生物技术通报, 2024, 40(8): 221-231. |
| [10] | 崔原瑗, 王昭懿, 白双宇, 任毓昭, 豆飞飞, 刘彩霞, 刘凤楼, 王掌军, 李清峰. 大麦非特异性磷脂酶C基因家族全基因组鉴定及苗期胁迫表达分析[J]. 生物技术通报, 2024, 40(8): 74-82. |
| [11] | 杨巍, 赵丽芬, 唐兵, 周麟笔, 杨娟, 莫传园, 张宝会, 李飞, 阮松林, 邓英. 芥菜SRO基因家族全基因组鉴定与表达分析[J]. 生物技术通报, 2024, 40(8): 129-141. |
| [12] | 周冉, 王兴平, 李彦霞, 罗仍卓么. 金黄色葡萄球菌型乳房炎奶牛乳腺组织的lncRNA差异表达分析[J]. 生物技术通报, 2024, 40(8): 320-328. |
| [13] | 李雨晴, 吴楠, 罗建让. 卵叶牡丹花色苷合成相关基因bHLH的克隆与功能分析[J]. 生物技术通报, 2024, 40(8): 174-185. |
| [14] | 张明亚, 庞胜群, 刘玉东, 苏永峰, 牛博文, 韩琼琼. 番茄FAD基因家族的鉴定与表达分析[J]. 生物技术通报, 2024, 40(7): 150-162. |
| [15] | 臧文蕊, 马明, 砗根, 哈斯阿古拉. 甜瓜BZR转录因子家族基因的全基因组鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(7): 163-171. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||