








生物技术通报 ›› 2025, Vol. 41 ›› Issue (7): 172-180.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0043
黄丹丹1( ), 吴云翼1, 邹建华1, 俞婷1, 朱炎辉2, 杨梅宏1, 董文丽1, 高冬丽1(
), 吴云翼1, 邹建华1, 俞婷1, 朱炎辉2, 杨梅宏1, 董文丽1, 高冬丽1( )
)
收稿日期:2025-01-12
出版日期:2025-07-26
发布日期:2025-07-22
通讯作者:
高冬丽,女,研究员,研究方向 :马铃薯淀粉调控;E-mail: gdongli@126.com作者简介:黄丹丹,硕士研究生,研究方向 :马铃薯基因功能解析;E-mail: yeee_hkx@163.com
基金资助:
HUANG Dan-dan1( ), WU Yun-yi1, ZOU Jian-hua1, YU Ting1, ZHU Yan-hui2, YANG Mei-hong1, DONG Wen-li1, GAO Dong-li1(
), WU Yun-yi1, ZOU Jian-hua1, YU Ting1, ZHU Yan-hui2, YANG Mei-hong1, DONG Wen-li1, GAO Dong-li1( )
)
Received:2025-01-12
Published:2025-07-26
Online:2025-07-22
摘要:
目的 PROTEIN TARGETING TO STARCH(PTST)在淀粉合成中发挥重要功能,本研究旨在探究马铃薯(Solanum tuberosum L.)StPTST2a基因的作用。 方法 利用反转录技术克隆StPTST2a基因,并对其氨基酸序列进行生物信息分析;利用荧光定量PCR技术分析StPTST2a在不同组织的表达量;利用原核表达系统在体外表达融合MBP标签的StPTST2a蛋白,并对融合蛋白进行体外淀粉结合实验;克隆7个淀粉合成相关基因,利用酵母双杂交及荧光素酶互补实验揭示StPTST2a与淀粉合成相关基因之间的互作关系。 结果 StPTST2a基因编码了一个具有526个氨基酸的蛋白,在C端含有一个CBM48结构域。该基因在各个组织均有表达,其中在叶片中的表达较高。体外淀粉结合实验表明,StPTST2a是一个淀粉结合蛋白。分析StPTST2a与7个淀粉合成相关基因的相互作用关系,发现StPTST2a与StSS4、StSS6和StISA1.1在酵母和植物体内均有互作。 结论 StPTST2a能够与多个淀粉合成基因形成复合体,在马铃薯淀粉合成和淀粉颗粒形成中发挥潜在作用。
黄丹丹, 吴云翼, 邹建华, 俞婷, 朱炎辉, 杨梅宏, 董文丽, 高冬丽. 马铃薯StPTST2a基因的克隆及互作分析[J]. 生物技术通报, 2025, 41(7): 172-180.
HUANG Dan-dan, WU Yun-yi, ZOU Jian-hua, YU Ting, ZHU Yan-hui, YANG Mei-hong, DONG Wen-li, GAO Dong-li. Cloning and Interaction Analysis of StPTST2a Gene in Potato[J]. Biotechnology Bulletin, 2025, 41(7): 172-180.
| 引物名称Primer name | 引物序列 Primer sequence (5′-3′) | 引物用途 Primer usage |
|---|---|---|
| pMAL-C2X-StPTST2a-F | GGATCGAGGGAAGGATTTCAATGGTTTTGACCCAGTTCGC | 体外淀粉结合实验 |
| pMAL-C2X-StPTST2a-R | CAGGTCGACTCTAGAGGATCCTTATGTGACAATGAGAAGATT | In vitro starch-binding assays |
| pGBKT7-StPTST2a-F | CATATGGCCATGGAGGCCATGGTTTTGACCCAGTTCG | 酵母双杂交实验 |
| pGBKT7-StPTST2a-R | CTAGTTATGCGGCCGCTGCAGTTATGTGACAATGAGAAG | Yeast two-hybrid assays |
| pGADT7-StSS1-F | CATATGGCCATGGAGGCCAGTATGGGGTCTCTGCAAACAC | 酵母双杂交实验 |
| pGADT7-StSS1-R | CGATGCCCACCCGGGTGTCATCTGACATATGGAGGATC | Yeast two-hybrid assays |
| pGADT7-StSS2-F | CATATGGCCATGGAGGCCAGTATGGAGAATTCCATTCTTC | 酵母双杂交实验 |
| pGADT7-StSS2-R | CGATGCCCACCCGGGTGTCACCACTGATACTTAGCAG | Yeast two-hybrid assays |
| pGADT7-StSS3-F | CATATGGCCATGGAGGCCAGTATGGACTCACTTACAGTTCT | 酵母双杂交实验 |
| pGADT7-StSS3-R | CGATGCCCACCCGGGTGCTATTCTAACTTTCTAGCAGC | Yeast two-hybrid assays |
| pGADT7-StSS4-F | 酵母双杂交实验 | |
| pGADT7-StSS4-R | Yeast two-hybrid assays | |
| pGADT7-StSS5-F | 酵母双杂交实验 | |
| pGADT7-StSS5-R | Yeast two-hybrid assays | |
| pGADT7-StSS6-F | CATGGAGGCCAGTATGGATTTCACATCCGGCCTGTC | 酵母双杂交实验 |
| pGADT7-StSS6-R | CGATGCCCACCCGGGTGTCAAAGCAAGTGACGTCTAAG | Yeast two-hybrid assays |
| pGADT7-StISA1.1-F | ATGGCCATGGAGGCCAGTATGGTACGGCAGTTCATCAA | 酵母双杂交实验 |
| pGADT7-StISA1.1-R | CGATGCCCACCCGGGTGCTATGCATCATCAGCAGATG | Yeast two-hybrid assays |
| pCAMBIA-nLUC-StPTST2a-F | GAACAGGGGGACGAGCTCATGGTTTTGACCCAGTTCG | 荧光素酶互补实验 |
| pCAMBIA-nLUC-StPTST2a-R | CCCGGGACGCGTACGAGATCTGTGTGACAATGAGAAGATTA | Luciferase complementation assays |
| pCAMBIA-cLUC-StSS4-F | CTCGTACGCGTCCCGGGGCATGGAGATGAAGATCTCC | 荧光素酶互补实验 |
| pCAMBIA-cLUC-StSS4-R | CGTCCTTGTAGTCCATTTGTTTCAACTACGACTTGCAGCTCTTGC | Luciferase complementation assays |
| pCAMBIA-cLUC-StSS6-F | CTCGTACGCGTCCCGGGGCATGGATTTCACATCCGG | 荧光素酶互补实验 |
| pCAMBIA-cLUC-StSS6-R | GTCCTTGTAGTCCATTTGTTTCAAAGCAAGTGACGTCTAAGTAC | Luciferase complementation assays |
| pCAMBIA-cLUC-StISA1.1-F | GTACGCGTCCCGGGGCATGGTACGGCAGTTCAT | 荧光素酶互补实验 |
| pCAMBIA-cLUC-StISA1.1-R | CGTCCTTGTAGTCCATTTGTTCTATGCATCATCAGCAGATGATAG | Luciferase complementation assays |
| qPCR-StPTST2a-F | CCAGCTTCCTCATCTGGTAGATC | 荧光定量PCR |
| qPCR-StPTST2a-R | CATGTCGTGAGGCTTCCATTTTT | Quantitative real-time PCR |
表1 分子实验所用引物
Table 1 Primers used for molecular experiments
| 引物名称Primer name | 引物序列 Primer sequence (5′-3′) | 引物用途 Primer usage |
|---|---|---|
| pMAL-C2X-StPTST2a-F | GGATCGAGGGAAGGATTTCAATGGTTTTGACCCAGTTCGC | 体外淀粉结合实验 |
| pMAL-C2X-StPTST2a-R | CAGGTCGACTCTAGAGGATCCTTATGTGACAATGAGAAGATT | In vitro starch-binding assays |
| pGBKT7-StPTST2a-F | CATATGGCCATGGAGGCCATGGTTTTGACCCAGTTCG | 酵母双杂交实验 |
| pGBKT7-StPTST2a-R | CTAGTTATGCGGCCGCTGCAGTTATGTGACAATGAGAAG | Yeast two-hybrid assays |
| pGADT7-StSS1-F | CATATGGCCATGGAGGCCAGTATGGGGTCTCTGCAAACAC | 酵母双杂交实验 |
| pGADT7-StSS1-R | CGATGCCCACCCGGGTGTCATCTGACATATGGAGGATC | Yeast two-hybrid assays |
| pGADT7-StSS2-F | CATATGGCCATGGAGGCCAGTATGGAGAATTCCATTCTTC | 酵母双杂交实验 |
| pGADT7-StSS2-R | CGATGCCCACCCGGGTGTCACCACTGATACTTAGCAG | Yeast two-hybrid assays |
| pGADT7-StSS3-F | CATATGGCCATGGAGGCCAGTATGGACTCACTTACAGTTCT | 酵母双杂交实验 |
| pGADT7-StSS3-R | CGATGCCCACCCGGGTGCTATTCTAACTTTCTAGCAGC | Yeast two-hybrid assays |
| pGADT7-StSS4-F | 酵母双杂交实验 | |
| pGADT7-StSS4-R | Yeast two-hybrid assays | |
| pGADT7-StSS5-F | 酵母双杂交实验 | |
| pGADT7-StSS5-R | Yeast two-hybrid assays | |
| pGADT7-StSS6-F | CATGGAGGCCAGTATGGATTTCACATCCGGCCTGTC | 酵母双杂交实验 |
| pGADT7-StSS6-R | CGATGCCCACCCGGGTGTCAAAGCAAGTGACGTCTAAG | Yeast two-hybrid assays |
| pGADT7-StISA1.1-F | ATGGCCATGGAGGCCAGTATGGTACGGCAGTTCATCAA | 酵母双杂交实验 |
| pGADT7-StISA1.1-R | CGATGCCCACCCGGGTGCTATGCATCATCAGCAGATG | Yeast two-hybrid assays |
| pCAMBIA-nLUC-StPTST2a-F | GAACAGGGGGACGAGCTCATGGTTTTGACCCAGTTCG | 荧光素酶互补实验 |
| pCAMBIA-nLUC-StPTST2a-R | CCCGGGACGCGTACGAGATCTGTGTGACAATGAGAAGATTA | Luciferase complementation assays |
| pCAMBIA-cLUC-StSS4-F | CTCGTACGCGTCCCGGGGCATGGAGATGAAGATCTCC | 荧光素酶互补实验 |
| pCAMBIA-cLUC-StSS4-R | CGTCCTTGTAGTCCATTTGTTTCAACTACGACTTGCAGCTCTTGC | Luciferase complementation assays |
| pCAMBIA-cLUC-StSS6-F | CTCGTACGCGTCCCGGGGCATGGATTTCACATCCGG | 荧光素酶互补实验 |
| pCAMBIA-cLUC-StSS6-R | GTCCTTGTAGTCCATTTGTTTCAAAGCAAGTGACGTCTAAGTAC | Luciferase complementation assays |
| pCAMBIA-cLUC-StISA1.1-F | GTACGCGTCCCGGGGCATGGTACGGCAGTTCAT | 荧光素酶互补实验 |
| pCAMBIA-cLUC-StISA1.1-R | CGTCCTTGTAGTCCATTTGTTCTATGCATCATCAGCAGATGATAG | Luciferase complementation assays |
| qPCR-StPTST2a-F | CCAGCTTCCTCATCTGGTAGATC | 荧光定量PCR |
| qPCR-StPTST2a-R | CATGTCGTGAGGCTTCCATTTTT | Quantitative real-time PCR |

图1 StPTST2a的序列分析、同源基因比对及表达模式分析A:StPTST2a和StPTST2b蛋白结构示意图;coiled-coil(CC)结构域用灰色标注,CBM结构域用蓝色标注,序列长度在上方标注;B:StPTST2a和其同源基因的氨基酸序列比对;StPTST2a的CC、CBM结构域在下方标注;C:StPTST2a在马铃薯CIP065各组织的表达水平
Fig. 1 Sequence analysis, homologous gene comparison and expression pattern analysis of StPTST2aA: A structural diagram of the StPTST2a and StPTST2b protein. The coiled-coil (CC) domain is marked in gray, the CBM domain is marked in blue, and the sequence length is labeled above. B: An amino acid sequence alignment of StPTST2a and its homologous genes. The CC and CBM domain of StPTST2a are labeled below. C: The transcription level of StPTST2a in various tissues of potato CIP065
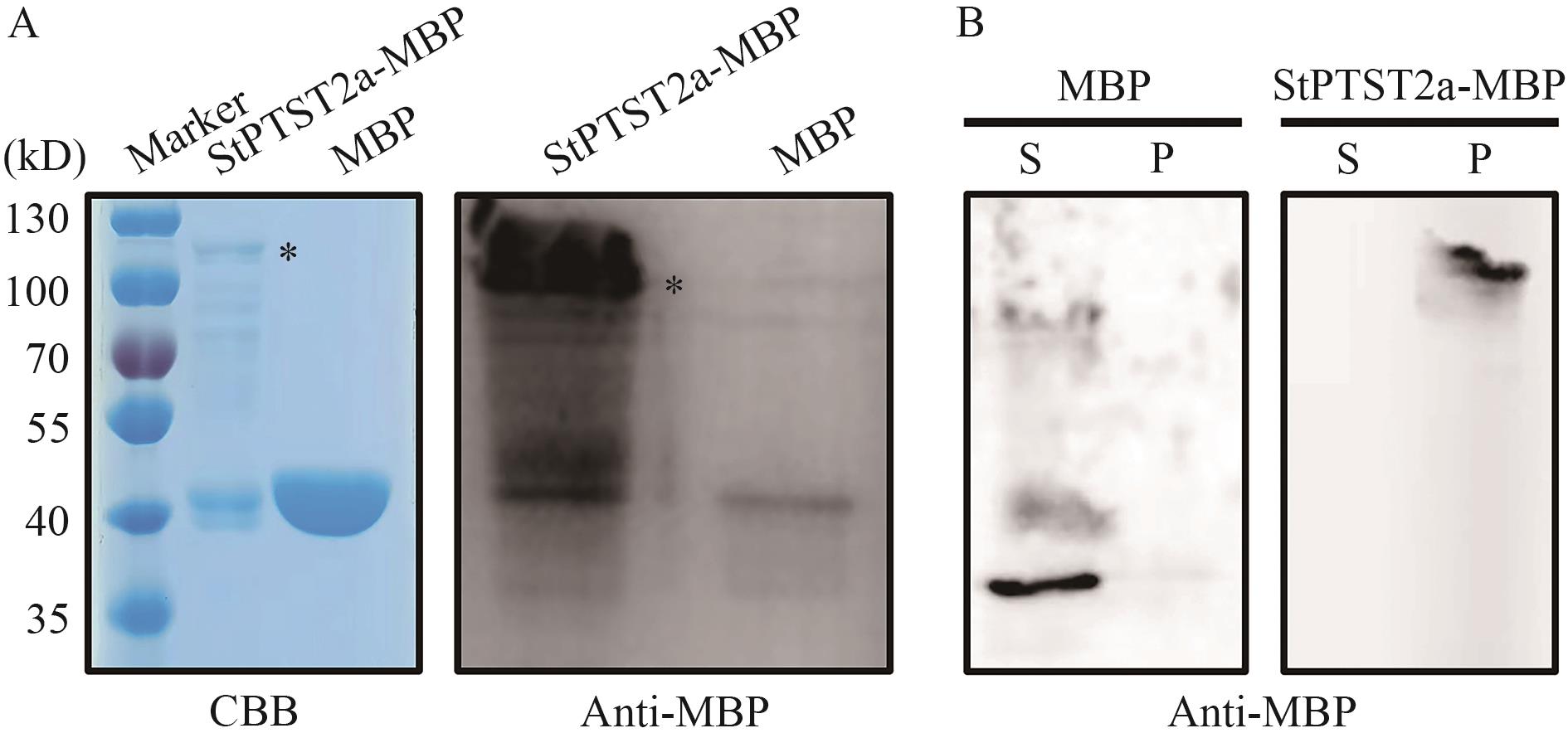
图2 StPTST2a蛋白在体外与淀粉结合A:纯化后融合蛋白StPTST2a-MBP与空MBP蛋白进行SDS-PAGE检测,并用考马斯亮蓝染色,星号显示融合蛋白StPTST2a-MBP。B:融合蛋白StPTST2a-MBP与空MBP蛋白分别与淀粉共孵育,每个样品的上清(S)和沉淀(P)均用MBP抗体进行检测
Fig. 2 StPTST2a protein binds to starch in vitroA: The recombinant fusion protein StPTST2a-MBP and empty MBP protein were subjected to SDS-PAGE detection and stained with Coomassie Brilliant Blue (CBB). The asterisk indicates the fusion protein StPTST2a-MBP. B: The recombinant fusion protein StPTST2a-MBP and empty MBP protein were incubated with starch separately, and both the supernatant (S) and pellet (P) from each sample were detected with an MBP antibody
基因 Gene | 基因号 Gene ID | 花 Flower | 叶 Leaf | 叶柄 Petiole | 茎 Stem | 匍匐茎Stolon | 发育块茎Young tuber | 成熟块茎 Mature tuber |
|---|---|---|---|---|---|---|---|---|
| StSS1 | PGSC0003DMG402018552 | 25.61 | 63.81 | 33.58 | 22.92 | 14.46 | 19.21 | 14.80 |
| StSS2 | PGSC0003DMG400001328 | 21.37 | 59.03 | 37.83 | 50.03 | 45.99 | 124.37 | 207.38 |
| StSS3 | PGSC0003DMG400016481 | 9.03 | 18.75 | 15.66 | 10.55 | 10.48 | 35.72 | 40.04 |
| StSS4 | PGSC0003DMG400008322 | 10.81 | 6.04 | 7.51 | 6.26 | 14.39 | 8.27 | 4.13 |
| StSS5 | PGSC0003DMG400030619 | 3.71 | 1.25 | 1.42 | 3.84 | 14.25 | 42.0 | 57.44 |
| StSS6 | PGSC0003DMG402013540 | 2.28 | 2.00 | 7.30 | 4.10 | 8.72 | 13.75 | 13.21 |
| StISA1.1 | PGSC0003DMG400020699 | 5.29 | 19.7 | 11.29 | 7.52 | 16.16 | 38.48 | 42.26 |
表2 StSS1-StSS6及StISA1.1基因在马铃薯RH各组织的FPKM值
Table 2 FPKM values of StSS1-StSS6 and StISA1.1 genes in different tissues of potato RH
基因 Gene | 基因号 Gene ID | 花 Flower | 叶 Leaf | 叶柄 Petiole | 茎 Stem | 匍匐茎Stolon | 发育块茎Young tuber | 成熟块茎 Mature tuber |
|---|---|---|---|---|---|---|---|---|
| StSS1 | PGSC0003DMG402018552 | 25.61 | 63.81 | 33.58 | 22.92 | 14.46 | 19.21 | 14.80 |
| StSS2 | PGSC0003DMG400001328 | 21.37 | 59.03 | 37.83 | 50.03 | 45.99 | 124.37 | 207.38 |
| StSS3 | PGSC0003DMG400016481 | 9.03 | 18.75 | 15.66 | 10.55 | 10.48 | 35.72 | 40.04 |
| StSS4 | PGSC0003DMG400008322 | 10.81 | 6.04 | 7.51 | 6.26 | 14.39 | 8.27 | 4.13 |
| StSS5 | PGSC0003DMG400030619 | 3.71 | 1.25 | 1.42 | 3.84 | 14.25 | 42.0 | 57.44 |
| StSS6 | PGSC0003DMG402013540 | 2.28 | 2.00 | 7.30 | 4.10 | 8.72 | 13.75 | 13.21 |
| StISA1.1 | PGSC0003DMG400020699 | 5.29 | 19.7 | 11.29 | 7.52 | 16.16 | 38.48 | 42.26 |

图4 StPTST2a与淀粉合成相关酶的互作分析A:StPTST2a与StSS1-StSS6、StISA1.1之间的酵母双杂交实验。B:StPTST2a与StSS4、StSS6、StISA1.1的荧光素酶互补实验
Fig. 4 Analysis of interaction between StPTST2a and enzymes related to starch synthesisA: Yeast two-hybrid assays between StPTST2a, StSS1-StSS6 and StISA1.1. B: Luciferase complementation assays of StPTST2a with StSS4, StSS6 and StISA1.1
| [1] | 赵宇慈, 许丹, 靳承煜, 等. 马铃薯块茎干物质、淀粉及还原糖含量的检测及相关性分析 [J]. 现代食品科技, 2017, 33(10): 288-293, 280. |
| Zhao YC, Xu D, Jin CY, et al. Detection and correlation analysis of dry matter, starch and reducing sugar content in potato tubers [J]. Mod Food Sci Technol, 2017, 33(10): 288-293, 280. | |
| [2] | Zeeman SC, Kossmann J, Smith AM. Starch: its metabolism, evolution, and biotechnological modification in plants [J]. Annu Rev Plant Biol, 2010, 61: 209-234. |
| [3] | 何虎翼, 唐洲萍, 杨鑫, 等. 马铃薯淀粉合成与降解研究进展 [J]. 生物技术通报, 2019, 35(4): 101-107. |
| He HY, Tang ZP, Yang X, et al. Research progress on potato starch synthesis and degradation [J]. Biotechnol Bull, 2019, 35(4): 101-107. | |
| [4] | Ohdan T, Francisco PB Jr, Sawada T, et al. Expression profiling of genes involved in starch synthesis in sink and source organs of rice [J]. J Exp Bot, 2005, 56(422): 3229-3244. |
| [5] | Liu HM, Yu GL, Wei B, et al. Identification and phylogenetic analysis of a novel starch synthase in maize [J]. Front Plant Sci, 2015, 6: 1013. |
| [6] | He S, Hao X, Wang S, et al. A newly-identified inactive starch synthase simultaneously regulates starch synthesis and carbon allocation in storage roots of cassava [J]. BioRxiv, 2020. |
| [7] | Cantarel BL, Coutinho PM, Rancurel C, et al. The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics [J]. Nucleic Acids Res, 2009, 37(Database issue): D233-D238. |
| [8] | Seung D, Soyk S, Coiro M, et al. Protein targeting to starch is required for localising granule-bound starch synthase to starch granules and for normal amylose synthesis in Arabidopsis [J]. PLoS Biol, 2015, 13(2): e1002080. |
| [9] | Wang W, Wei XJ, Jiao GA, et al. GBSS-BINDING PROTEIN, encoding a CBM48 domain-containing protein, affects rice quality and yield [J]. J Integr Plant Biol, 2020, 62(7): 948-966. |
| [10] | Seung D, Boudet J, Monroe J, et al. Homologs of protein targeting to starch control starch granule initiation in Arabidopsis leaves [J]. Plant Cell, 2017, 29(7): 1657-1677. |
| [11] | Seung D, Schreier TB, Bürgy L, et al. Two plastidial coiled-coil proteins are essential for normal starch granule initiation in Arabidopsis [J]. Plant Cell, 2018, 30(7): 1523-1542. |
| [12] | Peng C, Wang YH, Liu F, et al. FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm [J]. Plant J, 2014, 77(6): 917-930. |
| [13] | Zhang L, Li N, Zhang J, et al. The CBM48 domain-containing protein FLO6 regulates starch synthesis by interacting with SSIVb and GBSS in rice [J]. Plant Mol Biol, 2022, 108(4/5): 343-361. |
| [14] | Kerk D, Conley TR, Rodriguez FA, et al. A chloroplast-localized dual-specificity protein phosphatase in Arabidopsis contains a phylogenetically dispersed and ancient carbohydrate-binding domain, which binds the polysaccharide starch [J]. Plant J, 2006, 46(3): 400-413. |
| [15] | 石振明, 杨慧芹, 高冬丽. 马铃薯PTST1基因的克隆、表达及互作分析 [J]. 植物生理学报, 2023, 59(8): 1575-1582. |
| Shi ZM, Yang HQ, Gao DL. Cloning of PTST1 and analysis of its expression and interaction in potato [J]. Plant Physiol J, 2023, 59(8): 1575-1582. | |
| [16] | Hochmuth A, Carswell M, Rowland A, et al. Distinct effects of PTST2b and MRC on starch granule morphogenesis in potato tubers [J]. Plant Biotechnol J, 2025, 23(2): 412-429. |
| [17] | Van Harsselaar JK, Lorenz J, Senning M, et al. Genome-wide analysis of starch metabolism genes in potato (Solanum tuberosum L.) [J]. BMC Genomics, 2017, 18(1): 37. |
| [18] | Rahman S, Nakamura Y, Li Z, et al. The sugary-type isoamylase gene from rice and Aegilops tauschii: characterization and comparison with maize and Arabidopsis [J]. Genome, 2003, 46(3): 496-506. |
| [19] | Roldán I, Wattebled F, Mercedes Lucas M, et al. The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation [J]. Plant J, 2007, 49(3): 492-504. |
| [20] | Hawkins E, Chen JW, Watson-Lazowski A, et al. STARCH SYNTHASE 4 is required for normal starch granule initiation in amyloplasts of wheat endosperm [J]. New Phytol, 2021, 230(6): 2371-2386. |
| [21] | Streb S, Delatte T, Umhang M, et al. Starch granule biosynthesis in Arabidopsis is abolished by removal of all debranching enzymes but restored by the subsequent removal of an endoamylase [J]. Plant Cell, 2008, 20(12): 3448-3466. |
| [22] | Hussain H, Mant A, Seale R, et al. Three isoforms of isoamylase contribute different catalytic properties for the debranching of potato glucans [J]. Plant Cell, 2003, 15(1): 133-149. |
| [23] | Utsumi Y, Utsumi C, Sawada T, et al. Functional diversity of isoamylase oligomers: the ISA1 Homo-oligomer is essential for amylopectin biosynthesis in rice endosperm [J]. Plant Physiol, 2011, 156(1): 61-77. |
| [24] | Burton RA, Jenner H, Carrangis L, et al. Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity [J]. Plant J, 2002, 31(1): 97-112. |
| [25] | Ferreira SJ, Senning M, Fischer-Stettler M, et al. Simultaneous silencing of isoamylases ISA1, ISA2 and ISA3 by multi-target RNAi in potato tubers leads to decreased starch content and an early sprouting phenotype [J]. PLoS One, 2017, 12(7): e0181444. |
| [1] | 王从欢, 伍国强, 魏明. 植物CBL调控逆境胁迫响应的作用机制[J]. 生物技术通报, 2025, 41(7): 1-16. |
| [2] | 李霞, 张泽伟, 刘泽军, 王楠, 郭江波, 辛翠花, 张彤, 简磊. 马铃薯转录因子StMYB96的克隆及功能研究[J]. 生物技术通报, 2025, 41(7): 181-192. |
| [3] | 罗稷林, 栗锦烨, 贾玉鑫. 马铃薯中重力响应调节基因鉴定及功能分析[J]. 生物技术通报, 2025, 41(6): 109-118. |
| [4] | 许慧珍, SHANTWANA Ghimire, RAJU Kharel, 岳云, 司怀军, 唐勋. 马铃薯SUMO E3连接酶基因家族分析及StSIZ1基因的克隆与表达模式分析[J]. 生物技术通报, 2025, 41(6): 119-129. |
| [5] | 段永红, 杨欣, 于冠群, 夏俊俊, 宋陆帅, 白小东, 彭锁堂. 125份马铃薯种质资源遗传多样性及主成分分析[J]. 生物技术通报, 2025, 41(6): 130-143. |
| [6] | 宋慧洋, 苏宝杰, 李京昊, 梅超, 宋倩娜, 崔福柱, 冯瑞云. 马铃薯StAS2-15基因的克隆及盐胁迫下功能分析[J]. 生物技术通报, 2025, 41(5): 119-128. |
| [7] | 文博霖, 万敏, 胡建军, 王克秀, 景晟林, 王心悦, 朱博, 唐铭霞, 李兵, 何卫, 曾子贤. 马铃薯川芋50遗传转化及基因编辑体系的建立[J]. 生物技术通报, 2025, 41(4): 88-97. |
| [8] | 刘涛, 王志淇, 吴文博, 石文婷, 王超楠, 杜崇, 杨中敏. 马铃薯GRAM基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(4): 145-155. |
| [9] | 王涛, 胡社伟, 张宇, 邓文文, 尚春缘, 王婉艺. 玉米籽粒淀粉生物合成及调控因素研究进展[J]. 生物技术通报, 2025, 41(3): 1-13. |
| [10] | 张益瑄, 马宇, 王童童, 盛苏奥, 宋家凤, 吕钊彦, 朱晓彪, 侯华兰. 马铃薯DIR家族全基因组鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(3): 123-136. |
| [11] | 俞婷, 黄丹丹, 朱炎辉, 杨梅宏, 艾菊, 高冬丽. 马铃薯Stpatatin 05基因转录调控因子筛选及互作验证[J]. 生物技术通报, 2025, 41(3): 137-145. |
| [12] | 覃悦, 杨妍, 张磊, 卢丽丽, 李先平, 蒋伟. 二倍体和四倍体马铃薯StGAox基因鉴定与比较分析[J]. 生物技术通报, 2025, 41(3): 146-160. |
| [13] | 韩江涛, 张帅博, 秦雅蕊, 韩硕洋, 张雅康, 王吉庆, 杜清洁, 肖怀娟, 李猛. 甜瓜β-淀粉酶基因家族的鉴定及对非生物胁迫的响应[J]. 生物技术通报, 2025, 41(3): 171-180. |
| [14] | 杨涌, 袁国梅, 康肖肖, 刘亚明, 王东升, 张海娥. 板栗SWEET基因家族成员的鉴定及表达分析[J]. 生物技术通报, 2025, 41(2): 257-269. |
| [15] | 方慧敏, 顾艺枢, 张晶, 张龙. 水稻叶片淀粉的分离与理化性质分析[J]. 生物技术通报, 2025, 41(2): 51-57. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||