








生物技术通报 ›› 2025, Vol. 41 ›› Issue (3): 1-13.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0922
• 综述与专论 •
王涛1,2,3( ), 胡社伟1,2,3, 张宇1,2,3, 邓文文1, 尚春缘1, 王婉艺1,2(
), 胡社伟1,2,3, 张宇1,2,3, 邓文文1, 尚春缘1, 王婉艺1,2( )
)
收稿日期:2024-09-24
出版日期:2025-03-26
发布日期:2025-03-20
通讯作者:
王婉艺,女,博士,讲师,研究方向 :玉米遗传育种;E-mail: 2022110005@bzuu.edu.cn作者简介:王涛,男,博士,讲师,研究方向 :玉米籽粒营养物质代谢;E-mail: 2022110014@bzuu.edu.cn
基金资助:
WANG Tao1,2,3( ), HU She-wei1,2,3, ZHANG Yu1,2,3, DENG Wen-wen1, SHANG Chun-yuan1, WANG Wan-yi1,2(
), HU She-wei1,2,3, ZHANG Yu1,2,3, DENG Wen-wen1, SHANG Chun-yuan1, WANG Wan-yi1,2( )
)
Received:2024-09-24
Published:2025-03-26
Online:2025-03-20
摘要:
玉米作为一种在全球范围内广泛栽培的作物,不仅是人类饮食结构中不可或缺的粮食来源,而且在饲料产业及清洁能源领域同样发挥着重要作用。其籽粒中富含的淀粉,构成了玉米生物量的主体,直接决定了玉米的产量水平,是农业生产和食品加工中不可或缺的原料。然而,随着全球人口的不断增长、城市化进程的加速推进,以及气候变化带来的农业用地减少和自然灾害频发,全球粮食供应体系正面临着前所未有的挑战。玉米作为世界重要农作物之一,通过解析其淀粉生物合成途径培育高产优良品种变得十分重要。本文总结了参与玉米淀粉生物合成途径的关键酶及转运蛋白,并探讨这些因子在淀粉合成过程中的具体作用。同时,对合成途径中关键酶的突变体籽粒表型进行综述,分析基因突变对淀粉产量与品质的影响。此外,本文还总结了调控淀粉生物合成的各种因素,探讨如何通过优化这些因素来提高玉米的产量与品质。最后,本文展望了未来的研究方向,旨在为优质高产玉米的分子设计育种提供坚实的理论基础与实践指导。
王涛, 胡社伟, 张宇, 邓文文, 尚春缘, 王婉艺. 玉米籽粒淀粉生物合成及调控因素研究进展[J]. 生物技术通报, 2025, 41(3): 1-13.
WANG Tao, HU She-wei, ZHANG Yu, DENG Wen-wen, SHANG Chun-yuan, WANG Wan-yi. Research Progress in Starch Biosynthesis and Regulatory Factors in Maize Kernel[J]. Biotechnology Bulletin, 2025, 41(3): 1-13.
| 酶/转运蛋白 | 同工酶 | 基因名称 | 基因号 | 玉米突变体表型 |
|---|---|---|---|---|
| Enzymes/Transport proteins | Isoenzymes | Gene name | Gene ID | Phenotypes of maize mutants |
| 蔗糖合成酶 | SUS-sh1 | Shrunken1(Sh1) | Zm00001d045042 | 籽粒冠部塌陷,淀粉含量显著降低[ |
| SUS1 | Sus1 | Zm00001d047253 | 籽粒无明显缺陷表型、淀粉含量无显著变化,新鲜籽粒蔗糖含量累积[ | |
| SUS2 | Sus2 | Zm00001d029091 | 与sus1表型基本一致[ | |
| ADP-葡萄糖焦磷酸化酶 | AGPL3 | Shrunken2 (Sh2)/Zmagpl3 | Zm00001d044129 | 籽粒淀粉含量显著下降、蔗糖水平上升,籽粒胚乳严重皱缩、粒重降低[ |
| AGPL2 | Agplemzm/ZmagpL2 | Zm00001d005546 | / | |
| AGPL1 | Agpllzm/ZmagpL1 | Zm00001d019266 | 叶片淀粉含量显著下降;籽粒无明显缺陷表型、粒重略微降低[ | |
| AGPS2 | Brittle2 (Bt2)/Zmagps2 | Zm00001d050032 | 籽粒淀粉含量显著下降、蔗糖水平上升,籽粒胚乳严重皱缩、质地酥脆易碎、粒重降低[ | |
| AGPS3 | Agpsemzm/Zmagps3 | Zm00001d039131 | 籽粒胚淀粉含量显著下降、胚乳淀粉略微下降,籽粒无明显缺陷表型、粒重略微降低[ | |
| AGPS1 | Agpslzm/Zmagps1 | Zm00001d032385 | 叶片淀粉含量显著下降、蔗糖含量上升;籽粒无明显缺陷表型、粒重略微降低[ | |
| ADPG转运蛋白 | Brittle1 | Brittle1 (Bt1) | Zm00001d015746 | 籽粒胚乳中淀粉含量严重下降,籽粒皱缩、多棱、粒重显著降低[ |
| 淀粉合成酶 | GBSSI | Waxy (Wx) | Zm00001d045462 | 籽粒支链淀粉几乎占总淀粉含量100%,籽粒胚乳不透明、无光泽、外观晦暗呈蜡质状[ |
| GBSSIIa | ZmGBSSIIa | Zm00001d019479 | / | |
| GBSSIIb | ZmGBSSIIb | Zm00001d027242 | / | |
| SSI | SSI | Zm00001d045261 | / | |
| SSIIa | Sugary2 (Su2) | Zm00001d037234 | 籽粒淀粉含量下降、直链淀粉上升10%-15%,籽粒胚乳半透明、呈玻璃状、无光泽[ | |
| SSIII | Dull1 (Du1) | Zm00001d000002 | 根据遗传背景的不同籽粒直链淀粉含量略微或显著上升,籽粒胚乳呈玻璃状、无光泽、外观钝化[ | |
| 淀粉分支酶 | SBEI | Sbe1 | Zm00001d014844 | 籽粒和叶片淀粉性质和含量几乎没有变化[ |
| SBEIIa | Sbe2a | Zm00001d011301 | 叶片早期衰老,叶片淀粉累积、淀粉分支数减少、分支长度增加;籽粒淀粉性质和含量几乎没有变化[ | |
| SBEIIb | Amylose Extender (Ae) | Zm00001d016684 | 籽粒直链淀粉含量由20%上升至50%,籽粒胚乳透明呈玻璃状、体积变小、种皮皱缩[ | |
| 去淀粉分支酶 | ISA1 | Sugary1 (Su1) | Zm00001d049753 | 籽粒总淀粉和支链淀粉降低、植物糖原、多种单糖以及蔗糖含量积累,籽粒外观皱缩凹陷[ |
| ISA2 | Sugary4 (Su4) | Zm00001d038121 | 籽粒总淀粉与野生型基本一致,籽粒无明显缺陷表型[ | |
| ISA3 | ISO3 | Zm00001d020799 | / | |
| PUL | Zpu1 | Zm00001d004438 | 叶片和籽粒中淀粉分解代谢受阻,籽粒中淀粉性质和含量几乎没有变化,籽粒无明显缺陷表型[ | |
| 淀粉磷酸化酶 | PHOL/PHO1 | Shrunken4 (Sh4) | Zm00001d034074 | 籽粒总淀粉含量显著降低,籽粒胚乳皱缩且粉质[ |
| PHOH/PHO2 | ZmPHOH/Chl3 | Zm00001d042842 | 叶片细胞叶绿体中的瞬时淀粉累积,叶片褪绿、光合效率显著降低[ |
表1 参与玉米淀粉生物合成的酶和非酶蛋白因子
Table 1 Enzymes and non-enzymatic proteins involved in maize starch biosynthesis
| 酶/转运蛋白 | 同工酶 | 基因名称 | 基因号 | 玉米突变体表型 |
|---|---|---|---|---|
| Enzymes/Transport proteins | Isoenzymes | Gene name | Gene ID | Phenotypes of maize mutants |
| 蔗糖合成酶 | SUS-sh1 | Shrunken1(Sh1) | Zm00001d045042 | 籽粒冠部塌陷,淀粉含量显著降低[ |
| SUS1 | Sus1 | Zm00001d047253 | 籽粒无明显缺陷表型、淀粉含量无显著变化,新鲜籽粒蔗糖含量累积[ | |
| SUS2 | Sus2 | Zm00001d029091 | 与sus1表型基本一致[ | |
| ADP-葡萄糖焦磷酸化酶 | AGPL3 | Shrunken2 (Sh2)/Zmagpl3 | Zm00001d044129 | 籽粒淀粉含量显著下降、蔗糖水平上升,籽粒胚乳严重皱缩、粒重降低[ |
| AGPL2 | Agplemzm/ZmagpL2 | Zm00001d005546 | / | |
| AGPL1 | Agpllzm/ZmagpL1 | Zm00001d019266 | 叶片淀粉含量显著下降;籽粒无明显缺陷表型、粒重略微降低[ | |
| AGPS2 | Brittle2 (Bt2)/Zmagps2 | Zm00001d050032 | 籽粒淀粉含量显著下降、蔗糖水平上升,籽粒胚乳严重皱缩、质地酥脆易碎、粒重降低[ | |
| AGPS3 | Agpsemzm/Zmagps3 | Zm00001d039131 | 籽粒胚淀粉含量显著下降、胚乳淀粉略微下降,籽粒无明显缺陷表型、粒重略微降低[ | |
| AGPS1 | Agpslzm/Zmagps1 | Zm00001d032385 | 叶片淀粉含量显著下降、蔗糖含量上升;籽粒无明显缺陷表型、粒重略微降低[ | |
| ADPG转运蛋白 | Brittle1 | Brittle1 (Bt1) | Zm00001d015746 | 籽粒胚乳中淀粉含量严重下降,籽粒皱缩、多棱、粒重显著降低[ |
| 淀粉合成酶 | GBSSI | Waxy (Wx) | Zm00001d045462 | 籽粒支链淀粉几乎占总淀粉含量100%,籽粒胚乳不透明、无光泽、外观晦暗呈蜡质状[ |
| GBSSIIa | ZmGBSSIIa | Zm00001d019479 | / | |
| GBSSIIb | ZmGBSSIIb | Zm00001d027242 | / | |
| SSI | SSI | Zm00001d045261 | / | |
| SSIIa | Sugary2 (Su2) | Zm00001d037234 | 籽粒淀粉含量下降、直链淀粉上升10%-15%,籽粒胚乳半透明、呈玻璃状、无光泽[ | |
| SSIII | Dull1 (Du1) | Zm00001d000002 | 根据遗传背景的不同籽粒直链淀粉含量略微或显著上升,籽粒胚乳呈玻璃状、无光泽、外观钝化[ | |
| 淀粉分支酶 | SBEI | Sbe1 | Zm00001d014844 | 籽粒和叶片淀粉性质和含量几乎没有变化[ |
| SBEIIa | Sbe2a | Zm00001d011301 | 叶片早期衰老,叶片淀粉累积、淀粉分支数减少、分支长度增加;籽粒淀粉性质和含量几乎没有变化[ | |
| SBEIIb | Amylose Extender (Ae) | Zm00001d016684 | 籽粒直链淀粉含量由20%上升至50%,籽粒胚乳透明呈玻璃状、体积变小、种皮皱缩[ | |
| 去淀粉分支酶 | ISA1 | Sugary1 (Su1) | Zm00001d049753 | 籽粒总淀粉和支链淀粉降低、植物糖原、多种单糖以及蔗糖含量积累,籽粒外观皱缩凹陷[ |
| ISA2 | Sugary4 (Su4) | Zm00001d038121 | 籽粒总淀粉与野生型基本一致,籽粒无明显缺陷表型[ | |
| ISA3 | ISO3 | Zm00001d020799 | / | |
| PUL | Zpu1 | Zm00001d004438 | 叶片和籽粒中淀粉分解代谢受阻,籽粒中淀粉性质和含量几乎没有变化,籽粒无明显缺陷表型[ | |
| 淀粉磷酸化酶 | PHOL/PHO1 | Shrunken4 (Sh4) | Zm00001d034074 | 籽粒总淀粉含量显著降低,籽粒胚乳皱缩且粉质[ |
| PHOH/PHO2 | ZmPHOH/Chl3 | Zm00001d042842 | 叶片细胞叶绿体中的瞬时淀粉累积,叶片褪绿、光合效率显著降低[ |
| 基因名称 | 基因号 | 转录因子家族 | 靶基因/潜在靶基因 | 启动子核心元件 | 参考文献 |
|---|---|---|---|---|---|
| Gene name | Gene ID | Transcription factor family | Target genes/Potential target genes | The core element of promoter | References |
| ZmThx20 | Zm00001d017420 | GT-2 trihelix | PHOH, SBEIV, GBSSI, Isoamylase1 | / | [ |
| ZmTCP7 | Zm00001d010730 | TCP | Bt2, Bt1, SSI, SSIIa, SSIIIa, Sh2, SSIV | GAACCCCAC | [ |
| ZmDof3 | Zm00001d035651 | DOF | Du1, Su2 | AAAG | [ |
| PBF | Zm00001d005100 | DOF | PPDK1, PPDK2, SSIII, SSIIa, SBE | AAAG | [ |
| ZmDOF36 | Zm00001d029512 | DOF | AGPS1a, AGPL1, GBSSI, SSIIa, ISA1, ISA3 | / | [ |
| ZmMADS1a | Zm00001d041781 | MADS | AGPL1, AGPS1a, SBEIIb, GBSSI, ISA1, SSIIa | / | [ |
| ZmES22 | Zm00001d031620 | MADS | SBEIIa, SBEIIb, GBSSI, GBSSII, AGPS, AGPL, ISA1, ISA2, ISA3, PUL, SSI, SSIIa, SSIIIa, SSIVa | / | [ |
| ZmNAC126 | Zm00001d005028 | NAC | GBSSI, SSIIa, SSIV, ISA1, ISA2, Sh2, BT2, GBSSI, SSIIIa, BT1 | CACG | [ |
| ZmNAC34/ZmNAC128 | Zm00001d040189 | NAC | Bt2, Zpu1, SSI, SUS1, GBSSI, SBEIIb | ACGCAA | [ |
| ZmNAC130 | Zm00001d008403 | NAC | Bt2, Zpu1, SSI, SUS1, GBSSI, SBEIIb | ACGCAA | [ |
| ZmaNAC36 | Zm00001d003052 | NAC | AGPL2, AGPS2, SSI, GBSSIIb, SBEI | TTGCGTGTT | [ |
| ZmABI4 | Zm00001d038001 | AP2/EREBP | SSI | CACCG | [ |
| ZmEREB156 | Zm00001d026447 | AP2/EREBP | Sh2, SSIIIa | / | [ |
| ZmEREB94 | Zm00001d020043 | AP2/EREBP | SSI, Sh2, GBSSI, Bt1, SBEI, SSIIa, SSIV, ISA1 | / | [ |
| ZmPLATZ2 | Zm00001d029437 | PLATZ | SSI, SSIIa, SSIIIa, SSIV, SSV, ISA1, ISA2, Sh2, Bt2, GBSSI | CAAAAAAA | [ |
| ZmMYB14 | Zm00001d021537 | MYB | BT1, SSI, SSII, SBEI, GBSSI, Sh2, Bt2, ISA1 | TAACTG | [ |
| ZmMYB138 | Zm00001d043131 | MYB | BT2, Ae1, SSIIa | / | [ |
| ZmMYB115 | Zm00001d000361 | MYB | Du1, Wx | / | [ |
| ZmABI19 | Zm00001d011712 | B3 domain | O2, PBF, ZmbZIP22, ZmNAC130, O11 | TGCATG | [ |
| ZmbZIP29 | Zm00001d034571 | bZIP | O2 | / | [ |
| mEmBP-1 | Zm00001d019166 | bZIP | Sbe1 | / | [ |
| ZmbZIP91/ZmbZIP22 | Zm00001d021191 | bZIP | AGPS1, SSI, SSIIIa, ISA1, AGPL1a, SBEIIa, GBSSIa, ISA2, PHOL, SSIa | ACTCAT | [ |
| O2 | Zm00001d018971 | bZIP | PPDK1, PPDK2, SSIII, Sh1, SUS1, SUS2 | ACGT | [ |
| O11 | Zm00001d003677 | bHLH | O2, PBF, ZmDof3 | CACGTG | [ |
| ZmICE1 | Zm00001d042263 | bHLH | SSIIa, GBSSI | CANNTG | [ |
| NKD1 | Zm00001d002654 | IDD | O2 | TTGTCGT | [ |
| NKD2 | Zm00001d026113 | IDD | O2 | / | [ |
表2 玉米中参与调控淀粉生物合成的转录因子
Table 2 Transcription factors involved in regulating starch biosynthesis in maize
| 基因名称 | 基因号 | 转录因子家族 | 靶基因/潜在靶基因 | 启动子核心元件 | 参考文献 |
|---|---|---|---|---|---|
| Gene name | Gene ID | Transcription factor family | Target genes/Potential target genes | The core element of promoter | References |
| ZmThx20 | Zm00001d017420 | GT-2 trihelix | PHOH, SBEIV, GBSSI, Isoamylase1 | / | [ |
| ZmTCP7 | Zm00001d010730 | TCP | Bt2, Bt1, SSI, SSIIa, SSIIIa, Sh2, SSIV | GAACCCCAC | [ |
| ZmDof3 | Zm00001d035651 | DOF | Du1, Su2 | AAAG | [ |
| PBF | Zm00001d005100 | DOF | PPDK1, PPDK2, SSIII, SSIIa, SBE | AAAG | [ |
| ZmDOF36 | Zm00001d029512 | DOF | AGPS1a, AGPL1, GBSSI, SSIIa, ISA1, ISA3 | / | [ |
| ZmMADS1a | Zm00001d041781 | MADS | AGPL1, AGPS1a, SBEIIb, GBSSI, ISA1, SSIIa | / | [ |
| ZmES22 | Zm00001d031620 | MADS | SBEIIa, SBEIIb, GBSSI, GBSSII, AGPS, AGPL, ISA1, ISA2, ISA3, PUL, SSI, SSIIa, SSIIIa, SSIVa | / | [ |
| ZmNAC126 | Zm00001d005028 | NAC | GBSSI, SSIIa, SSIV, ISA1, ISA2, Sh2, BT2, GBSSI, SSIIIa, BT1 | CACG | [ |
| ZmNAC34/ZmNAC128 | Zm00001d040189 | NAC | Bt2, Zpu1, SSI, SUS1, GBSSI, SBEIIb | ACGCAA | [ |
| ZmNAC130 | Zm00001d008403 | NAC | Bt2, Zpu1, SSI, SUS1, GBSSI, SBEIIb | ACGCAA | [ |
| ZmaNAC36 | Zm00001d003052 | NAC | AGPL2, AGPS2, SSI, GBSSIIb, SBEI | TTGCGTGTT | [ |
| ZmABI4 | Zm00001d038001 | AP2/EREBP | SSI | CACCG | [ |
| ZmEREB156 | Zm00001d026447 | AP2/EREBP | Sh2, SSIIIa | / | [ |
| ZmEREB94 | Zm00001d020043 | AP2/EREBP | SSI, Sh2, GBSSI, Bt1, SBEI, SSIIa, SSIV, ISA1 | / | [ |
| ZmPLATZ2 | Zm00001d029437 | PLATZ | SSI, SSIIa, SSIIIa, SSIV, SSV, ISA1, ISA2, Sh2, Bt2, GBSSI | CAAAAAAA | [ |
| ZmMYB14 | Zm00001d021537 | MYB | BT1, SSI, SSII, SBEI, GBSSI, Sh2, Bt2, ISA1 | TAACTG | [ |
| ZmMYB138 | Zm00001d043131 | MYB | BT2, Ae1, SSIIa | / | [ |
| ZmMYB115 | Zm00001d000361 | MYB | Du1, Wx | / | [ |
| ZmABI19 | Zm00001d011712 | B3 domain | O2, PBF, ZmbZIP22, ZmNAC130, O11 | TGCATG | [ |
| ZmbZIP29 | Zm00001d034571 | bZIP | O2 | / | [ |
| mEmBP-1 | Zm00001d019166 | bZIP | Sbe1 | / | [ |
| ZmbZIP91/ZmbZIP22 | Zm00001d021191 | bZIP | AGPS1, SSI, SSIIIa, ISA1, AGPL1a, SBEIIa, GBSSIa, ISA2, PHOL, SSIa | ACTCAT | [ |
| O2 | Zm00001d018971 | bZIP | PPDK1, PPDK2, SSIII, Sh1, SUS1, SUS2 | ACGT | [ |
| O11 | Zm00001d003677 | bHLH | O2, PBF, ZmDof3 | CACGTG | [ |
| ZmICE1 | Zm00001d042263 | bHLH | SSIIa, GBSSI | CANNTG | [ |
| NKD1 | Zm00001d002654 | IDD | O2 | TTGTCGT | [ |
| NKD2 | Zm00001d026113 | IDD | O2 | / | [ |
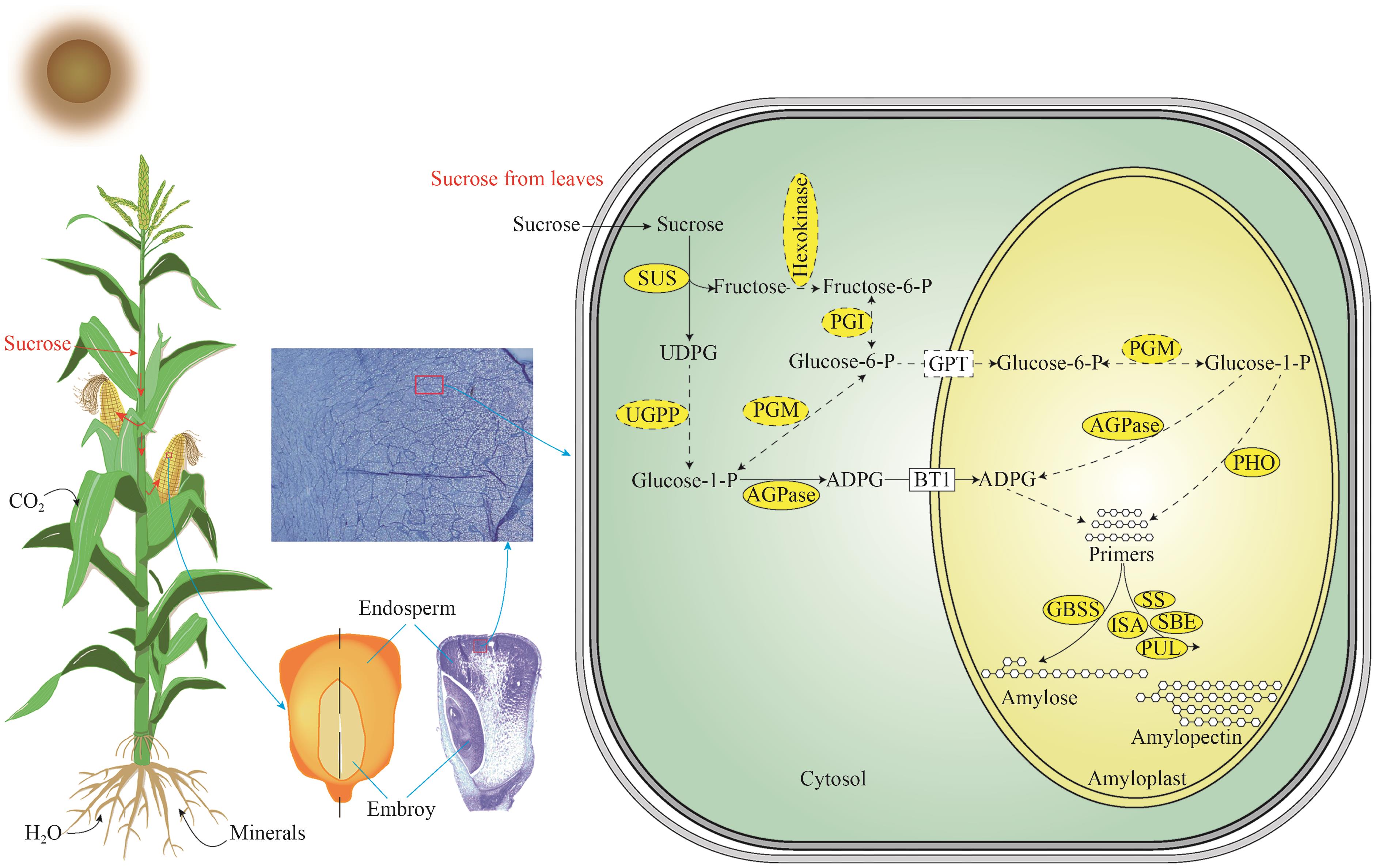
图1 玉米胚乳淀粉生物合成路径图中黑色箭头显示了玉米胚乳淀粉常规的生物合成步骤,虚线箭头代表推测的淀粉生物合成步骤,椭圆代表玉米中已被发现的淀粉生物合成路径关键酶,虚线椭圆代表玉米中未被验证功能的酶
Fig. 1 The pathway of starch biosynthesis in maize endospermThe black arrows show conventional steps of starch biosynthesis in maize endosperm, dashed arrows indicate the predicted steps of starch biosynthesis, the ellipses indicate the critical enzymes of starch biosynthesis pathway that have been discovered in maize, the dashed ellipses indicate the enzymes in maize whose function have not been verified
| 1 | Nuss ET, Tanumihardjo SA. Maize: a paramount staple crop in the context of global nutrition [J]. Compr Rev Food Sci Food Saf, 2010, 9(4): 417-436. |
| 2 | Nuss ET, Tanumihardjo SA. Quality protein maize for Africa: closing the protein inadequacy gap in vulnerable populations [J]. Adv Nutr, 2011, 2(3): 217-224. |
| 3 | Dai DW, Ma ZY, Song RT. Maize kernel development [J]. Mol Breed, 2021, 41(1): 2-9. |
| 4 | Hu ST, Wang M, Zhang X, et al. Genetic basis of kernel starch content decoded in a maize multi-parent population [J]. Plant Biotechnol J, 2021, 19(11): 2192-2205. |
| 5 | Zhu JH, Yu WW, Zhang CQ, et al. New insights into amylose and amylopectin biosynthesis in rice endosperm [J]. Carbohydr Polym, 2020, 230: 115656. |
| 6 | Finegan C, Boehlein SK, Leach KA, et al. Genetic perturbation of the starch biosynthesis in maize endosperm reveals sugar-responsive gene networks [J]. Front Plant Sci, 2022, 12: 800326. |
| 7 | Li RQ, Tan YY, Zhang HL. Regulators of starch biosynthesis in cereal crops [J]. Molecules, 2021, 26(23): 7092. |
| 8 | Huang LC, Tan HY, Zhang CQ, et al. Starch biosynthesis in cereal endosperms: an updated review over the last decade [J]. Plant Commun, 2021, 2(5): 100237. |
| 9 | 徐妙云, 邢利娟, 杨明雨, 等. 高直链淀粉禾谷类作物种质创新与利用研究进展 [J]. 生物技术通报, 2022, 38(4): 20-28. |
| Xu MY, Xing LJ, Yang MY, et al. Research progress in germplasm innovation and utilization of high amylose cereal crops [J]. Biotechnol Bull, 2022, 38(4): 20-28. | |
| 10 | Deng YT, Wang JC, Zhang ZY, et al. Transactivation of Sus1 and Sus2 by Opaque2 is an essential supplement to sucrose synthase-mediated endosperm filling in maize [J]. Plant Biotechnol J, 2020, 18(9): 1897-1907. |
| 11 | Carlson SJ, Chourey PS, Helentjaris T, et al. Gene expression studies on developing kernels of maize sucrose synthase (SuSy) mutants show evidence for a third SuSy gene [J]. Plant Mol Biol, 2002, 49(1): 15-29. |
| 12 | Tsai CY, Nelson OE. Starch-deficient maize mutant lacking adenosine dephosphate glucose pyrophosphorylase activity [J]. Science, 1966, 151(3708): 341-343. |
| 13 | Huang BQ, Hennen-Bierwagen TA, Myers AM. Functions of multiple genes encoding ADP-glucose pyrophosphorylase subunits in maize endosperm, embryo, and leaf [J]. Plant Physiol, 2014, 164(2): 596-611. |
| 14 | Teas HJ, Teas AN. Heritable characters in maize [J]. J Hered, 1953, 44(4): 156-158. |
| 15 | Slewinski TL, Ma Y, Baker RF, et al. Determining the role of Tie-dyed1 in starch metabolism: epistasis analysis with a maize ADP-glucose pyrophosphorylase mutant lacking leaf starch [J]. J Hered, 2008, 99(6): 661-666. |
| 16 | Mangelsdorf PC. The genetics and morphology of some endosperm characters in maize [D]. Massachusetts: Harvard University, 1925. |
| 17 | Wentz JB. Heritable characters of maize [J]. J Hered, 1926, 17(9): 327-329. |
| 18 | Tsai CY. The function of the waxy locus in starch synthesis in maize endosperm [J]. Biochem Genet, 1974, 11(2): 83-96. |
| 19 | Zhang XL, Colleoni C, Ratushna V, et al. Molecular characterization demonstrates that the Zea mays gene sugary2 codes for the starch synthase isoform SSIIa [J]. Plant Mol Biol, 2004, 54(6): 865-879. |
| 20 | Li J, Corke H. Physicochemical properties of maize starches expressing dull and sugary-2 mutants in different genetic backgrounds [J]. J Agric Food Chem, 1999, 47(12): 4939-4943. |
| 21 | Davis JH, Kramer HH, Whistler RL. Expression of the gene du in the endosperm of maize [J]. Agron J, 1955, 47(5): 232-235. |
| 22 | Mangelsdorf PC. The inheritance of amylaceous sugary endosperm and its derivatives in maize [J]. Genetics, 1947, 32(5): 448-458. |
| 23 | Goren A, Ashlock D, Tetlow IJ. Starch formation inside plastids of higher plants [J]. Protoplasma, 2018, 255(6): 1855-1876. |
| 24 | Blauth SL, Kim KN, Klucinec JD, et al. Identification of Mutator insertional mutants of starch-branching enzyme1 (sbe1) in Zea mays L [J]. Plant Mol Biol, 2002, 48(3): 287-297. |
| 25 | Blauth SL, Yao Y, Klucinec JD, et al. Identification of mutator insertional mutants of starch-branching enzyme 2a in corn [J]. Plant Physiology, 2001, 125(3): 1396-1405. |
| 26 | Vineyard ML, Bear RP, MacMasters MM, et al. Development of “amylomaize” —corn hybrids with high amylose starch: I. genetic considerations [J]. Agron J, 1958, 50(10): 595-598. |
| 27 | 任红丽, 张军杰, 黄玉碧. 玉米ae基因的研究进展 [J]. 玉米科学, 2007, 15(5): 56-59. |
| Ren HL, Zhang JJ, Huang YB. Research progress of ae gene in maize [J]. J Maize Sci, 2007, 15(5): 56-59. | |
| 28 | Dinges JR, Colleoni C, Myers AM, et al. Molecular structure of three mutations at the maize sugary1 locus and their allele-specific phenotypic effects [J]. Plant Physiol, 2001, 125(3): 1406-1418. |
| 29 | James MG, Robertson DS, Myers AM. Characterization of the maize gene sugary1, a determinant of starch composition in kernels [J]. Plant Cell, 1995, 7(4): 417-429. |
| 30 | De Vries BD, Tracy WF. Characterization of endosperm carbohydrates in isa2-339 maize and interactions with su1-ref [J]. Crop Sci, 2016, 56(5): 2277-2286. |
| 31 | Lin QH, Huang BQ, Zhang MX, et al. Functional interactions between starch synthase III and isoamylase-type starch-debranching enzyme in maize endosperm[J]. Plant Physiol, 2012, 158(2): 679-692. |
| 32 | Dinges JR, Colleoni C, James MG, et al. Mutational analysis of the pullulanase-type debranching enzyme of maize indicates multiple functions in starch metabolism [J]. Plant Cell, 2003, 15(3): 666-680. |
| 33 | Winter H, Huber SC. Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes [J]. Crit Rev Biochem Mol Biol, 2000, 35(4): 253-289. |
| 34 | Koch K. Sucrose metabolism: regulatory mechanisms and pivotal roles in sugar sensing and plant development [J]. Curr Opin Plant Biol, 2004, 7(3): 235-246. |
| 35 | McCarty DR, Shaw JR, Hannah LC. The cloning, genetic mapping, and expression of the constitutive sucrose synthase locus of maize [J]. Proc Natl Acad Sci USA, 1986, 83(23): 9099-9103. |
| 36 | Duncan KA, Hardin SC, Huber SC. The three maize sucrose synthase isoforms differ in distribution, localization, and phosphorylation [J]. Plant Cell Physiol, 2006, 47(7): 959-971. |
| 37 | Shaw JR, Ferl RJ, Baier J, et al. Structural features of the maize sus1 gene and protein [J]. Plant Physiol, 1994, 106(4): 1659-1665. |
| 38 | 沙米西努尔·牙森, 杨淑楠, 李娟, 等. 玉米高直链淀粉合成通路研究进展及应用 [J]. 分子植物育种, 2024, 22(24): 8103-8109. |
| Shamshinur Y, Yang SN, Li J, et al. Research progress and application of high amylose synthesis pathway in maize [J]. Mol Plant Breed, 2024, 22(24): 8103-8109. | |
| 39 | Pfister B, Zeeman SC. Formation of starch in plant cells [J]. Cell Mol Life Sci, 2016, 73(14): 2781-2807. |
| 40 | Rösti S, Denyer K. Two paralogous genes encoding small subunits of ADP-glucose pyrophosphorylase in maize, Bt2 and L2, replace the single alternatively spliced gene found in other cereal species [J]. J Mol Evol, 2007, 65(3): 316-327. |
| 41 | Denyer K, Dunlap F, Thorbjørnsen T, et al. The major form of ADP-glucose pyrophosphorylase in maize endosperm is extra-plastidial [J]. Plant Physiol, 1996, 112(2): 779-785. |
| 42 | Huang BQ, Chen J, Zhang JJ, et al. Characterization of ADP-glucose pyrophosphorylase encoding genes in source and sink organs of maize [J]. Plant Mol Biol Report, 2011, 29(3): 563-572. |
| 43 | Sullivan TD, Kaneko Y. The maize brittle 1 gene encodes amyloplast membrane polypeptides [J]. Planta, 1995, 196(3): 477-484. |
| 44 | Kirchberger S, Leroch M, Huynen MA, et al. Molecular and biochemical analysis of the plastidic ADP-glucose transporter (ZmBT1) from Zea mays [J]. J Biol Chem, 2007, 282(31): 22481-22491. |
| 45 | Kötting O, Kossmann J, Zeeman SC, et al. Regulation of starch metabolism: the age of enlightenment? [J]. Curr Opin Plant Biol, 2010, 13(3): 321-329. |
| 46 | Crofts N, Abe N, Oitome NF, et al. Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes [J]. J Exp Bot, 2015, 66(15): 4469-4482. |
| 47 | Ohdan T, Francisco PB Jr, Sawada T, et al. Expression profiling of genes involved in starch synthesis in sink and source organs of rice [J]. J Exp Bot, 2005, 56(422): 3229-3244. |
| 48 | Yan HB, Pan XX, Jiang HW, et al. Comparison of the starch synthesis genes between maize and rice: copies, chromosome location and expression divergence [J]. Theor Appl Genet, 2009, 119(5): 815-825. |
| 49 | Irshad A, Guo HJ, Rehman SU, et al. Soluble starch synthase enzymes in cereals: an updated review [J]. Agronomy, 2021, 11(10): 1983. |
| 50 | Abt MR, Zeeman SC. Evolutionary innovations in starch metabolism [J]. Curr Opin Plant Biol, 2020, 55: 109-117. |
| 51 | Liu HM, Yu GL, Wei B, et al. Identification and phylogenetic analysis of a novel starch synthase in maize [J]. Front Plant Sci, 2015, 6: 1013. |
| 52 | Takeda Y, Preiss J. Structures of B90 (sugary) and W64A (normal) maize starches [J]. Carbohydr Res, 1993, 240: 265-275. |
| 53 | Shannon JC, Garwood DL. Genetics and physiology of starch development [M]//Starch: Chemistry and Technology. Amsterdam: Elsevier, 1984: 25-86. |
| 54 | Creech RG. Genetic control of carbohydrate synthesis in maize endosperm [J]. Genetics, 1965, 52(6): 1175-1186. |
| 55 | Cao H, James MG, Myers AM. Purification and characterization of soluble starch synthases from maize endosperm [J]. Arch Biochem Biophys, 2000, 373(1): 135-146. |
| 56 | Jespersen HM, MacGregor EA, Henrissat B, et al. Starch- and glycogen-debranching and branching enzymes: prediction of structural features of the catalytic (beta/alpha)8-barrel domain and evolutionary relationship to other amylolytic enzymes [J]. J Protein Chem, 1993, 12(6): 791-805. |
| 57 | Sawada T, Itoh M, Nakamura Y. Contributions of three starch branching enzyme isozymes to the fine structure of amylopectin in rice endosperm [J]. Front Plant Sci, 2018, 9: 1536. |
| 58 | Tetlow IJ, Emes MJ. A review of starch-branching enzymes and their role in amylopectin biosynthesis [J]. IUBMB Life, 2014, 66(8): 546-558. |
| 59 | Fisher DK, Gao M, Kim KN, et al. Allelic analysis of the maize amylose-extender locus suggests that independent genes encode starch-branching enzymes IIa and IIb [J]. Plant Physiol, 1996, 110(2): 611-619. |
| 60 | Brust H, Lehmann T, D'Hulst C, et al. Analysis of the functional interaction of Arabidopsis starch synthase and branching enzyme isoforms reveals that the cooperative action of SSI and BEs results in glucans with polymodal chain length distribution similar to amylopectin [J]. PLoS One, 2014, 9(7): e102364. |
| 61 | Liu FS, Romanova N, Lee EA, et al. Glucan affinity of starch synthase IIa determines binding of starch synthase I and starch-branching enzyme IIb to starch granules [J]. Biochem J, 2012, 448(3): 373-387. |
| 62 | Garwood DL, Shannon JC, Creech RG. Starches of endosperms possessing different alleles at the amylose extender locus in Zea mays L. [maize] [J]. Cereal Chemistry, 1976, 53(3): 355-364. |
| 63 | Gao M, Fisher DK, Kim KN, et al. Independent genetic control of maize starch-branching enzymes IIa and IIb. Isolation and characterization of a Sbe2a cDNA [J]. Plant Physiol, 1997, 114(1): 69-78. |
| 64 | Preiss J. Biology and molecular biology of starch synthesis and its regulation [M]//Oxford Surveys Of Plant Molecular And Cell Biology. Oxford University PressOxford, 1991,7: 59-114. |
| 65 | Guan HP, Preiss J. Differentiation of the properties of the branching isozymes from maize (Zea mays) [J]. Plant Physiol, 1993, 102(4): 1269-1273. |
| 66 | Takeda Y, Guan HP, Preiss J. Branching of amylose by the branching isoenzymes of maize endosperm [J]. Carbohydr Res, 1993, 240: 253-263. |
| 67 | Gao M, Fisher DK, Kim KN, et al. Evolutionary conservation and expression patterns of maize starch branching enzyme I and IIb genes suggests isoform specialization [J]. Plant Mol Biol, 1996, 30(6): 1223-1232. |
| 68 | Gyawali A, Auger D. Evidence that the allelic segregation of starch branching enzyme 1 (sbe1) is the source of a high amylose QTL in maize [J]. Crop Sci, 2018, 58(1): 98-102. |
| 69 | Kubo A, Colleoni C, Dinges JR, et al. Functions of heteromeric and homomeric isoamylase-type starch-debranching enzymes in developing maize endosperm [J]. Plant Physiol, 2010, 153(3): 956-969. |
| 70 | Yang RC, Yan ZG, Wang QF, et al. Marker-assisted backcrossing of lcyE for enhancement of ProA in sweet corn [J]. Euphytica, 2018, 214(8): 130. |
| 71 | Hossain F, Nepolean T, Vishwakarma AK, et al. Mapping and validation of microsatellite markers linked to sugary1 and shrunken2 genes in maize (Zea mays L.) [J]. J Plant Biochem Biotechnol, 2015, 24(2): 135-142. |
| 72 | Denyer K, Sidebottom C, Hylton CM, et al. Soluble isoforms of starch synthase and starch-branching enzyme also occur within starch granules in developing pea embryos [J]. Plant J, 1993, 4(1): 191-198. |
| 73 | Mu-Forster C, Huang R, Powers JR, et al. Physical association of starch biosynthetic enzymes with starch granules of maize endosperm. Granule-associated forms of starch synthase I and starch branching enzyme II [J]. Plant Physiol, 1996, 111(3): 821-829. |
| 74 | Denyer K, Waite D, Motawia S, et al. Granule-bound starch synthase I in isolated starch granules elongates malto-oligosaccharides processively [J]. Biochem J, 1999, 340 ( Pt 1)(Pt 1): 183-191. |
| 75 | Takeda Y, Maruta N, Hizukuri S. Structures of amylose subfractions with different molecular sizes [J]. Carbohydr Res, 1992, 226(2): 279-285. |
| 76 | Hanashiro I, Itoh K, Kuratomi Y, et al. Granule-bound starch synthase I is responsible for biosynthesis of extra-long unit chains of amylopectin in rice [J]. Plant Cell Physiol, 2008, 49(6): 925-933. |
| 77 | Zhang ZY, Zheng XX, Yang J, et al. Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis [J]. Proc Natl Acad Sci USA, 2016, 113(39): 10842-10847. |
| 78 | Dong Q, Xu QQ, Kong JJ, et al. Overexpression of ZmbZIP22 gene alters endosperm starch content and composition in maize and rice [J]. Plant Sci, 2019, 283: 407-415. |
| 79 | Yanagisawa S. Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants [J]. Plant Cell Physiol, 2004, 45(4): 386-391. |
| 80 | Yanagisawa S, Schmidt RJ. Diversity and similarity among recognition sequences of Dof transcription factors [J]. Plant J, 1999, 17(2): 209-214. |
| 81 | Olsen AN, Ernst HA, Leggio LL, et al. DNA-binding specificity and molecular functions of NAC transcription factors [J]. Plant Sci, 2005, 169(4): 785-797. |
| 82 | Zhang ZY, Dong JQ, Ji C, et al. NAC-type transcription factors regulate accumulation of starch and protein in maize seeds [J]. Proc Natl Acad Sci USA, 2019, 116(23): 11223-11228. |
| 83 | Peng XJ, Wang QQ, Wang Y, et al. A maize NAC transcription factor, ZmNAC34, negatively regulates starch synthesis in rice [J]. Plant Cell Rep, 2019, 38(12): 1473-1484. |
| 84 | Chen EW, Yu HQ, He J, et al. The transcription factors ZmNAC128 and ZmNAC130 coordinate with Opaque2 to promote endosperm filling in maize [J]. Plant Cell, 2023, 35(11): 4066-4090. |
| 85 | Zhang JJ, Chen J, Yi Q, et al. Novel role of ZmaNAC36 in co-expression of starch synthetic genes in maize endosperm [J]. Plant Mol Biol, 2014, 84(3): 359-369. |
| 86 | Xiao QL, Wang YY, Du J, et al. ZmMYB14 is an important transcription factor involved in the regulation of the activity of the ZmBT1 promoter in starch biosynthesis in maize [J]. FEBS J, 2017, 284(18): 3079-3099. |
| 87 | Feng F, Qi WW, Lv YD, et al. OPAQUE11 is a central hub of the regulatory network for maize endosperm development and nutrient metabolism [J]. Plant Cell, 2018, 30(2): 375-396. |
| 88 | Yang T, Guo LX, Ji C, et al. The B3 domain-containing transcription factor ZmABI19 coordinates expression of key factors required for maize seed development and grain filling [J]. Plant Cell, 2021, 33(1): 104-128. |
| 89 | Hu YF, Li YP, Weng JF, et al. Coordinated regulation of starch synthesis in maize endosperm by microRNAs and DNA methylation [J]. Plant J, 2021, 105(1): 108-123. |
| 90 | Li P, Li ZX, Xie GN, et al. Trihelix transcription factor ZmThx20 is required for kernel development in maize [J]. Int J Mol Sci, 2021, 22(22): 12137. |
| 91 | Ajayo BS, Li YP, Wang YY, et al. The novel ZmTCP7 transcription factor targets AGPase-encoding gene ZmBt2 to regulate storage starch accumulation in maize [J]. Front Plant Sci, 2022, 13: 943050. |
| 92 | Qi X, Li SX, Zhu YX, et al. ZmDof3, a maize endosperm-specific Dof protein gene, regulates starch accumulation and aleurone development in maize endosperm [J]. Plant Mol Biol, 2017, 93(1-2): 7-20. |
| 93 | Wu JD, Chen L, Chen MC, et al. The DOF-domain transcription factor ZmDOF36 positively regulates starch synthesis in transgenic maize [J]. Front Plant Sci, 2019, 10: 465. |
| 94 | Dong Q, Wang F, Kong JJ, et al. Functional analysis of ZmMADS1a reveals its role in regulating starch biosynthesis in maize endosperm [J]. Sci Rep, 2019, 9(1): 3253. |
| 95 | Zha KY, Xie HX, Ge M, et al. Expression of maize MADS transcription factor ZmES22 negatively modulates starch accumulation in rice endosperm [J]. Int J Mol Sci, 2019, 20(3): 483. |
| 96 | Xiao QL, Wang YY, Li H, et al. Transcription factor ZmNAC126 plays an important role in transcriptional regulation of maize starch synthesis-related genes [J]. Crop J, 2021, 9(1): 192-203. |
| 97 | Hu YF, Li YP, Zhang JJ, et al. Binding of ABI4 to a CACCG motif mediates the ABA-induced expression of the ZmSSI gene in maize (Zea mays L.) endosperm [J]. J Exp Bot, 2012, 63(16): 5979-5989. |
| 98 | Huang HH, Xie SD, Xiao QL, et al. Sucrose and ABA regulate starch biosynthesis in maize through a novel transcription factor, ZmEREB156 [J]. Sci Rep, 2016, 6: 27590. |
| 99 | Li H, Xiao QL, Zhang CX, et al. Identification and characterization of transcription factor ZmEREB94 involved in starch synthesis in maize [J]. J Plant Physiol, 2017, 216: 11-16. |
| 100 | Li H, Wang YY, Xiao QL, et al. Transcription factor ZmPLATZ2 positively regulate the starch synthesis in maize [J]. Plant Growth Regul, 2021, 93(3): 291-302. |
| 101 | Yang T, Wang HN, Guo LX, et al. Aba-induced phosphorylation of basic leucine zipper 29, abscisic acid insensitive 19, and Opaque2 by SnRK2.2 enhances gene transactivation for endosperm filling in maize [J]. Plant Cell, 2022, 34(5): 1933-1956. |
| 102 | Kim KN, Guiltinan MJ. Identification of cis-acting elements important for expression of the starch-branching enzyme I gene in maize endosperm [J]. Plant Physiol, 1999, 121(1): 225-236. |
| 103 | Li CB, Yue YH, Chen HJ, et al. The ZmbZIP22 transcription factor regulates 27-kD γ-zein gene transcription during maize endosperm development [J]. Plant Cell, 2018, 30(10): 2402-2424. |
| 104 | Liu HM, Wang YB, Liu LJ, et al. Pleiotropic ZmICE1 is an important transcriptional regulator of maize endosperm starch biosynthesis [J]. Front Plant Sci, 2022, 13: 895763. |
| 105 | Gontarek BC, Neelakandan AK, Wu H, et al. NKD transcription factors are central regulators of maize endosperm development [J]. Plant Cell, 2016, 28(12): 2916-2936. |
| 106 | Xie S, Zhang X, Zhou Z, et al. Identification of genes alternatively spliced in developing maize endosperm [J]. Plant Biol, 2018, 20(1): 59-66. |
| 107 | Varagona MJ, Purugganan M, Wessler SR. Alternative splicing induced by insertion of retrotransposons into the maize waxy gene [J]. Plant Cell, 1992, 4(7): 811-820. |
| 108 | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function [J]. Cell, 2004, 116(2): 281-297. |
| 109 | Li DD, Liu ZC, Gao L, et al. Genome-wide identification and characterization of microRNAs in developing grains of Zea mays L [J]. PLoS One, 2016, 11(4): e0153168. |
| 110 | Wang T, Chang YM, Zhao K, et al. Maize RNA 3′-terminal phosphate cyclase-like protein promotes 18S pre-rRNA cleavage and is important for kernel development [J]. Plant Cell, 2022, 34(5): 1957-1979. |
| 111 | Wu XL, Gong FP, Cao D, et al. Advances in crop proteomics: PTMs of proteins under abiotic stress [J]. Proteomics, 2016, 16(5): 847-865. |
| 112 | Walley JW, Shen ZX, Sartor R, et al. Reconstruction of protein networks from an atlas of maize seed proteotypes [J]. Proc Natl Acad Sci USA, 2013, 110(49): E4808-E4817. |
| 113 | Makhmoudova A, Williams D, Brewer D, et al. Identification of multiple phosphorylation sites on maize endosperm starch branching enzyme IIb, a key enzyme in amylopectin biosynthesis [J]. J Biol Chem, 2014, 289(13): 9233-9246. |
| 114 | Chen J, Huang BQ, Li YP, et al. Synergistic influence of sucrose and abscisic acid on the genes involved in starch synthesis in maize endosperm [J]. Carbohydr Res, 2011, 346(13): 1684-1691. |
| 115 | Hannah LC, Tuschall DM, Mans RJ. Multiple forms of maize endosperm adp-glucose pyrophosphorylase and their control by shrunken-2 and brittle-2 [J]. Genetics, 1980, 95(4): 961-970. |
| 116 | Boehlein SK, Liu P, Webster A, et al. Effects of long-term exposure to elevated temperature on Zea mays endosperm development during grain fill [J]. Plant J, 2019, 99(1): 23-40. |
| 117 | Cairns JE, Sonder K, Zaidi PH, et al. Maize production in a changing climate. impacts, adaptation, and mitigation strategies [J]. Adv Agron, 2012, 114: 1-58. |
| 118 | Guo J, Qu LL, Hu YF, et al. Proteomics reveals the effects of drought stress on the kernel development and starch formation of waxy maize [J]. BMC Plant Biol, 2021, 21(1): 434. |
| 119 | Yang H, Gu XT, Ding MQ, et al. Activities of starch synthetic enzymes and contents of endogenous hormones in waxy maize grains subjected to post-silking water deficit [J]. Sci Rep, 2019, 9(1): 7059. |
| 120 | Wu WH, Zhong YY, Liu YL, et al. A new insight into the biosynthesis, structure, and functionality of waxy maize starch under drought stress [J]. J Sci Food Agric, 2023, 103(11): 5270-5276. |
| 121 | Li CS, Huang YC, Huang RD, et al. The genetic architecture of amylose biosynthesis in maize kernel [J]. Plant Biotechnol J, 2018, 16(2): 688-695. |
| 122 | Liu N, Xue YD, Guo ZY, et al. Genome-wide association study identifies candidate genes for starch content regulation in maize kernels [J]. Front Plant Sci, 2016, 7: 1046. |
| 123 | Wu ZD, Wang ZQ, Zhang K. Isolation and functional characterization of a glucose-6-phosphate/phosphate translocator (IbG6PPT1) from sweet potato (Ipomoea batatas (L.) Lam.) [J]. BMC Plant Biol, 2021, 21(1): 595. |
| 124 | Qu AL, Xu Y, Yu XX, et al. Sporophytic control of anther development and male fertility by glucose-6-phosphate/phosphate translocator 1 (OsGPT1) in rice [J]. J Genet Genomics, 2021, 48(8): 695-705. |
| 125 | Flügge UL. Phosphate translocation in the regulation of photosynthesis [J]. J Exp Bot, 1995, 46: 1317-1323. |
| 126 | Kammerer B, Fischer K, Hilpert B, et al. Molecular characterization of a carbon transporter in plastids from heterotrophic tissues: the glucose 6-phosphate/phosphate antiporter [J]. Plant Cell, 1998, 10(1): 105-117. |
| 127 | Dong L, Qi XT, Zhu JJ, et al. Supersweet and waxy: meeting the diverse demands for specialty maize by genome editing [J]. Plant Biotechnol J, 2019, 17(10): 1853-1855. |
| 128 | Zhao YJ, Li N, Li B, et al. Reduced expression of starch branching enzyme IIa and IIb in maize endosperm by RNAi constructs greatly increases the amylose content in kernel with nearly normal morphology [J]. Planta, 2015, 241(2): 449-461. |
| 129 | Li PP, Ma HZ, Xiao N, et al. Overexpression of the ZmSUS1 gene alters the content and composition of endosperm starch in maize (Zea mays L.) [J]. Planta, 2023, 257(5): 97. |
| 130 | Singh J, Das S, Jagadis Gupta K, et al. Physiological implications of SWEETs in plants and their potential applications in improving source-sink relationships for enhanced yield [J]. Plant Biotechnol J, 2023, 21(8): 1528-1541. |
| [1] | 杨涌, 袁国梅, 康肖肖, 刘亚明, 王东升, 张海娥. 板栗SWEET基因家族成员的鉴定及表达分析[J]. 生物技术通报, 2025, 41(2): 257-269. |
| [2] | 方慧敏, 顾艺枢, 张晶, 张龙. 水稻叶片淀粉的分离与理化性质分析[J]. 生物技术通报, 2025, 41(2): 51-57. |
| [3] | 任晓敏, 云岚, 艾芊, 赵乔. 新麦草异戊烯基转移酶PjIPT基因的功能验证[J]. 生物技术通报, 2024, 40(7): 207-215. |
| [4] | 王秋月, 段鹏亮, 李海笑, 刘宁, 曹志艳, 董金皋. 玉米大斑病菌cDNA文库的构建及转录因子StMR1互作蛋白的筛选[J]. 生物技术通报, 2024, 40(6): 281-289. |
| [5] | 胡锦锦, 李素贞, 马旭辉, 柳小庆, 谢珊珊, 江海洋, 陈茹梅. 玉米花青素生物合成代谢调控[J]. 生物技术通报, 2024, 40(6): 34-44. |
| [6] | 孙亚楠, 王春雪, 王欣, 杜秉海, 刘凯, 汪城墙. 萎缩芽孢杆菌CNY01的生防特性及其对玉米的抗盐促生作用[J]. 生物技术通报, 2024, 40(5): 248-260. |
| [7] | 王佳玮, 李晨, 刘建利, 周世杰, 易嘉敏, 杨谨源, 康鹏. 内生真菌接种方式对青贮玉米幼苗生长的影响[J]. 生物技术通报, 2024, 40(4): 189-202. |
| [8] | 徐伟芳, 李贺宇, 张慧, 何仔昂, 高文恒, 谢紫洋, 王传文, 尹登科. 生防细菌HX0037对栝楼炭疽病的防病能力及其机制[J]. 生物技术通报, 2024, 40(4): 228-241. |
| [9] | 胡伊娃, 陈露. 玉米野生种基因组研究进展及应用[J]. 生物技术通报, 2024, 40(3): 14-24. |
| [10] | 李雪, 李容欧, 孔美懿, 黄磊. 解淀粉芽孢杆菌SQ-2对水稻的促生作用[J]. 生物技术通报, 2024, 40(2): 109-119. |
| [11] | 王楠, 廖永琴, 施竹凤, 申云鑫, 杨童雨, 冯路遥, 矣小鹏, 唐加菜, 陈齐斌, 杨佩文. 三株无量山森林土壤芽孢杆菌鉴定及其生物活性挖掘[J]. 生物技术通报, 2024, 40(2): 277-288. |
| [12] | 殷子薇, 红雨. 玫瑰红球菌NB1对玉米的耐盐促生效应及其全基因组研究[J]. 生物技术通报, 2024, 40(12): 193-207. |
| [13] | 王晶, 张晓磊, 白玉, 盛宇欣, 关海涛, 温洪涛. 不同玉米转化体通用PCR检测体系建立[J]. 生物技术通报, 2024, 40(12): 34-44. |
| [14] | 田锦, 张月秋, 张华, 陈子言, 田璐, 王颢潜, 高芳瑞, 梁晋刚, 陈红. 转基因玉米浙大瑞丰8特异性定性PCR检测方法研究[J]. 生物技术通报, 2024, 40(12): 45-52. |
| [15] | 戚旺, 毕一凡, 轩强兵, 张新, 周慧岗, 聂圆情, 程彪彪, 张禹舜, 王俊杰, 梁卫红. 水稻OsRhoGDI1过表达影响粒型及种子活力[J]. 生物技术通报, 2024, 40(11): 152-161. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||