








生物技术通报 ›› 2025, Vol. 41 ›› Issue (9): 289-301.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0215
• 研究报告 • 上一篇
李亚涛1( ), 张志鹏2,3, 赵梦瑶1, 吕镇1, 甘恬1, 魏浩3, 吴书凤3, 马玉超1(
), 张志鹏2,3, 赵梦瑶1, 吕镇1, 甘恬1, 魏浩3, 吴书凤3, 马玉超1( )
)
收稿日期:2025-03-02
出版日期:2025-09-26
发布日期:2025-09-24
通讯作者:
马玉超,女,副教授,研究方向 :微生物代谢工程;E-mail: mayuchao@bifu.edu.cn作者简介:李亚涛,男,硕士研究生,研究方向 :微生物代谢工程;E-mail: lyt888888@bjfu.edu.cn
基金资助:
LI Ya-tao1( ), ZHANG Zhi-peng2,3, ZHAO Meng-yao1, LYU Zhen1, GAN Tian1, WEI Hao3, WU Shu-feng3, MA Yu-chao1(
), ZHANG Zhi-peng2,3, ZHAO Meng-yao1, LYU Zhen1, GAN Tian1, WEI Hao3, WU Shu-feng3, MA Yu-chao1( )
)
Received:2025-03-02
Published:2025-09-26
Online:2025-09-24
摘要:
目的 从基因组水平了解大豆根瘤菌Bd1的遗传背景,探索促进Bd1生长和结瘤的有效途径。 方法 利用Illumina+PacBio三代测序平台对Bd1进行全基因组测序,采用生物信息学方法进行物种鉴定、功能基因注释、固氮相关基因和TetR家族转录因子分析,并利用同源重组双交换法构建TetR3基因敲除突变株,研究TetR3在Bd1生长和结瘤过程中的功能。 结果 Bd1与Bradyrhizobiumdiazoefficiens USDA110的基因组平均核苷酸一致性(ANI)和数字DNA-DNA杂交值分别为99.98%和98.7%。Bd1基因组大小为9 009 469 bp,平均GC含量为64.1%,共8 403个编码基因,基因平均长度为927 bp,含有51个tRNA编码基因和3套核糖体RNA基因;含共生结瘤固氮相关nod、nif和fix基因数量分别为9、12和10个;含结构多样的TetR家族转录因子编码基因58个。TetR3基因失活突变株增强了Bd1的生长和结瘤能力,且谷胱甘肽S-转移酶活性提高了41.0%。 结论 Bd1为B. diazoefficiens的1株新菌,具备与大豆共生结瘤固氮的能力,TetR3基因通过平衡细胞氧化还原能力负调控Bd1的生长和结瘤性能。
李亚涛, 张志鹏, 赵梦瑶, 吕镇, 甘恬, 魏浩, 吴书凤, 马玉超. 根瘤菌Bd1的全基因组分析及TetR3对细胞生长和结瘤的负调控功能[J]. 生物技术通报, 2025, 41(9): 289-301.
LI Ya-tao, ZHANG Zhi-peng, ZHAO Meng-yao, LYU Zhen, GAN Tian, WEI Hao, WU Shu-feng, MA Yu-chao. Whole Genome Analysis of Bradyrhizobium sp. Bd1 and the Negative Regulating Function of TetR3 during Cell Growth and Nodulation[J]. Biotechnology Bulletin, 2025, 41(9): 289-301.

图1 TetR3失活突变株的构建A:重组质粒和双亲结合供体的构建;B:TetR3失活结果;C:突变株菌株的测序结果;D:菌落PCR验证(M:DNA marker 2000;泳道1:Bd1;泳道2:Bd1ΔTetR3)
Fig. 1 Construction of TetR3-inactive mutantA: Construction of recombinant plasmid and conjugation donor. B: TetR3-inactive result. C: Sequencing of the mutant. D: Colony PCR verification (M: DNA marker 2000; lane 1: Bd1; lane 2: Bd1ΔTetR3)

图4 Bd1基因组的功能基因注释A:GOG功能注释;B:CAZy功能注释;C:GO功能注释;D:KEGG功能注释
Fig. 4 Functional gene annotation of Bd1 genomeA: GOG annotation; B: CAZy annotation; C: GO annotation; D: KEGG annotation
| Family | Gene ID | Name | KEGG gene ID | KO description | Location |
|---|---|---|---|---|---|
| fix | gene5832 | fixK | bja:bll3466 | CRP/FNR family transcriptional regulator | 6070103-6070786 |
| gene6542 | fixK | bja:bll2757 | CRP/FNR family transcriptional regulator | 6868843-6869541 | |
| gene6539 | fixL | bja:bll2760 | LuxR family, sensor kinase FixL | 6866141-6867658 | |
| gene6540 | fixJ | bja:bll2759 | LuxR family, response regulator FixJ | 6867662-6868279 | |
| gene7359 | fixA | bja:blr2038 | Flavoprotein beta subunit | 7803669-7804535 | |
| gene7895 | fixA | bja:blr1377 | Flavoprotein beta subunit | 8425108-8425857C | |
| gene7566 | fixX | bja:bsr1775 | Ferredoxin like protein | 8067228-8067524C | |
| gene7567 | fixC | bja:blr1774 | Flavoprotein-quinone oxidoreductase | 8067563-8068870C | |
| gene7568 | fixB | bja:blr1773 | Flavoprotein alpha subunit | 8068882-8069991C | |
| gene7894 | fixB | bja:blr1378 | Flavoprotein alpha subunit | 8424164-8425108C | |
| nod | gene5597 | nodT | bsym:CIT39_22430 | Multidrug efflux system | 5805752-5807308 |
| gene7363 | nodU | bja:blr2034 | Carbamoyltransferase | 7809897-7811504C | |
| gene7367 | nodU | bjp:RN69_37980 | Carbamoyltransferase | 7813893-7815602C | |
| gene7365 | nodJ | bja:blr2031 | Lipooligosaccharide transport system permease | 7812179-7812967C | |
| gene7366 | nodI | bja:blr2030 | Lipooligosaccharide transport system ATP-binding protein | 7812971-7813891C | |
| gene7368 | nodC | bja:blr2027 | N-acetylglucosaminyltransferase | 7816172-7817629C | |
| gene7369 | nodB | bja:blr2026 | Chitooligosaccharide deacetylase | 7817644-7818303C | |
| gene7370 | nodA | bja:blr2025 | Nodulation protein A | 7818300-7818932C | |
| gene7372 | nodD | bja:bll2023 | LysR family transcriptional regulator | 7819726-7820670 | |
| nif | gene7360 | nifA | bja:blr2037 | Nif-specific regulatory protein | 7805053-7806801C |
| gene7569 | nifW | bja:blr1771 | Nitrogenase-stabilizing/protective protein | 8070942-8071283C | |
| gene7571 | nifQ | bju:BJ6T_80490 | Nitrogen fixation protein NifQ | 8071938-8072714C | |
| gene7572 | nifH | bja:blr1769 | Nitrogenase iron protein NifH | 8072861-8073745 | |
| gene7580 | nifZ | bjp:RN69_39140 | Nitrogen fixation protein NifZ | 8078154-8078468C | |
| gene7582 | nifB | bju:BJ6T_80610 | Nitrogen fixation protein NifB | 8078954-8080537C | |
| gene7583 | nifT | bja:bsr1757 | Nitrogen fixation protein NifT | 8081201-8081425C | |
| gene7594 | nifX | bja:blr1747 | Nitrogen fixation protein NifX | 8090471-8090866C | |
| gene7595 | nifN | bja:blr1746 | Nitrogenase molybdenum-iron protein NifN | 8090863-8092272C | |
| gene7596 | nifE | bja:blr1745 | Nitrogenase molybdenum-cofactor synthesis protein NifE | 8092282-8093925C | |
| gene7597 | nifK | bja:blr1744 | Molybdenum-iron protein beta chain | 8094018-8095574C | |
| gene7598 | nifD | bja:blr1743 | Molybdenum-iron protein alpha chain | 8095640-8097142C |
表1 Bd1基因组中固氮相关基因
Table 1 Nitrogen fixation-related genes in Bd1 genome
| Family | Gene ID | Name | KEGG gene ID | KO description | Location |
|---|---|---|---|---|---|
| fix | gene5832 | fixK | bja:bll3466 | CRP/FNR family transcriptional regulator | 6070103-6070786 |
| gene6542 | fixK | bja:bll2757 | CRP/FNR family transcriptional regulator | 6868843-6869541 | |
| gene6539 | fixL | bja:bll2760 | LuxR family, sensor kinase FixL | 6866141-6867658 | |
| gene6540 | fixJ | bja:bll2759 | LuxR family, response regulator FixJ | 6867662-6868279 | |
| gene7359 | fixA | bja:blr2038 | Flavoprotein beta subunit | 7803669-7804535 | |
| gene7895 | fixA | bja:blr1377 | Flavoprotein beta subunit | 8425108-8425857C | |
| gene7566 | fixX | bja:bsr1775 | Ferredoxin like protein | 8067228-8067524C | |
| gene7567 | fixC | bja:blr1774 | Flavoprotein-quinone oxidoreductase | 8067563-8068870C | |
| gene7568 | fixB | bja:blr1773 | Flavoprotein alpha subunit | 8068882-8069991C | |
| gene7894 | fixB | bja:blr1378 | Flavoprotein alpha subunit | 8424164-8425108C | |
| nod | gene5597 | nodT | bsym:CIT39_22430 | Multidrug efflux system | 5805752-5807308 |
| gene7363 | nodU | bja:blr2034 | Carbamoyltransferase | 7809897-7811504C | |
| gene7367 | nodU | bjp:RN69_37980 | Carbamoyltransferase | 7813893-7815602C | |
| gene7365 | nodJ | bja:blr2031 | Lipooligosaccharide transport system permease | 7812179-7812967C | |
| gene7366 | nodI | bja:blr2030 | Lipooligosaccharide transport system ATP-binding protein | 7812971-7813891C | |
| gene7368 | nodC | bja:blr2027 | N-acetylglucosaminyltransferase | 7816172-7817629C | |
| gene7369 | nodB | bja:blr2026 | Chitooligosaccharide deacetylase | 7817644-7818303C | |
| gene7370 | nodA | bja:blr2025 | Nodulation protein A | 7818300-7818932C | |
| gene7372 | nodD | bja:bll2023 | LysR family transcriptional regulator | 7819726-7820670 | |
| nif | gene7360 | nifA | bja:blr2037 | Nif-specific regulatory protein | 7805053-7806801C |
| gene7569 | nifW | bja:blr1771 | Nitrogenase-stabilizing/protective protein | 8070942-8071283C | |
| gene7571 | nifQ | bju:BJ6T_80490 | Nitrogen fixation protein NifQ | 8071938-8072714C | |
| gene7572 | nifH | bja:blr1769 | Nitrogenase iron protein NifH | 8072861-8073745 | |
| gene7580 | nifZ | bjp:RN69_39140 | Nitrogen fixation protein NifZ | 8078154-8078468C | |
| gene7582 | nifB | bju:BJ6T_80610 | Nitrogen fixation protein NifB | 8078954-8080537C | |
| gene7583 | nifT | bja:bsr1757 | Nitrogen fixation protein NifT | 8081201-8081425C | |
| gene7594 | nifX | bja:blr1747 | Nitrogen fixation protein NifX | 8090471-8090866C | |
| gene7595 | nifN | bja:blr1746 | Nitrogenase molybdenum-iron protein NifN | 8090863-8092272C | |
| gene7596 | nifE | bja:blr1745 | Nitrogenase molybdenum-cofactor synthesis protein NifE | 8092282-8093925C | |
| gene7597 | nifK | bja:blr1744 | Molybdenum-iron protein beta chain | 8094018-8095574C | |
| gene7598 | nifD | bja:blr1743 | Molybdenum-iron protein alpha chain | 8095640-8097142C |
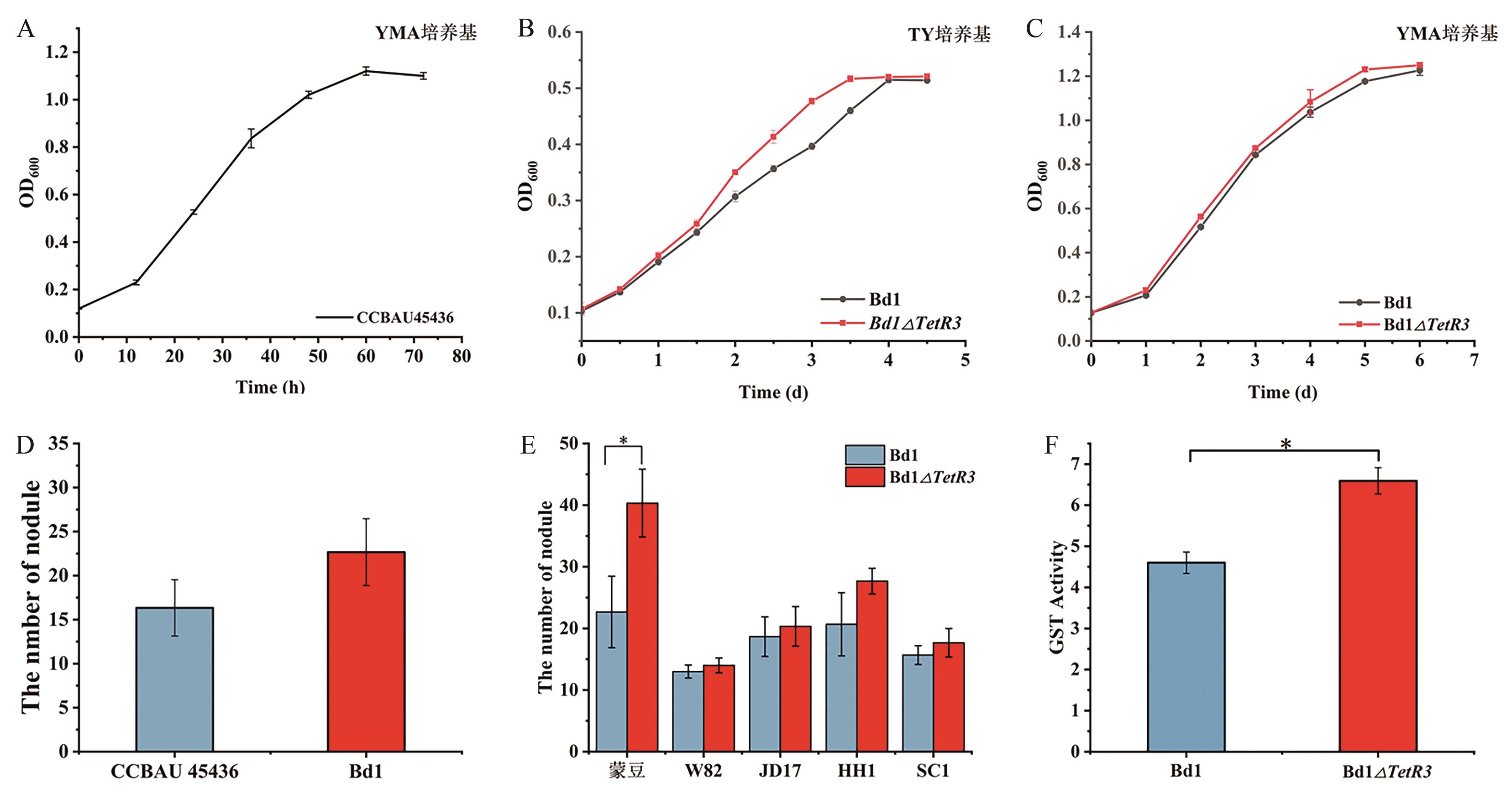
图7 Bd1和突变株Bd1ΔTetR3的功能特征A:快生根瘤菌CCBAU45436在YMA培养基中的生长曲线;B和C:突变株在YMA和TY培养基中的生长曲线;D:Bd1和快生根瘤菌(S. fredii CCBAU 45436)与蒙豆1137的结瘤数量;E:突变株Bd1ΔTetR3与不同品种大豆的结瘤数量;F:突变株Bd1ΔTetR3的谷胱甘肽S-转移酶活性。*P<0.05
Fig. 7 Functional characteristics of Bd1 and the mutant Bd1ΔTetR3A: Growth curve of S. fredii CCBAU45436 in YMA medium. B and C: Growth of the mutant Bd1ΔTetR3 in YMA and TY, respectively. D: Number of symbiotic nodules of Bd1 and S. fredii CCBAU 45436 with Glycine max Mengdou 1137; E: Number of symbiotic nodules between mutant and different soybean varieties. F: Glutathione S-transferase activity of the mutant Bd1ΔTetR3. *P<0.05
| [1] | 向星宇, 吴雪许, 许建双, 等. 中国大豆进口贸易现状及对策研究 [J]. 黑龙江粮食, 2024, (11): 26-28. |
| Xiang XY, Wu XX, Xu JS, et al. China's soybean import trade: current status and policy recommendations [J]. Heilongjiang Grain, 2024, (11): 26-28. | |
| [2] | 王善明. 华癸中慢生根瘤菌7653R全基因组测序及比较基因组学研究 [D]. 武汉: 华中农业大学, 2014. |
| Wang SM. Whole-genome sequencing and comparative genomic analysis of Mesorhizobium huakuii strain7653R [D]. Wuhan: Huazhong Agricultural University, 2014. | |
| [3] | Margaret I, Becker A, Blom J, et al. Symbiotic properties and first analyses of the genomic sequence of the fast growing model strain Sinorhizobium fredii HH103 nodulating soybean [J]. J Biotechnol, 2011, 155(1): 11-19. |
| [4] | Yang SH, Chen WH, Wang ET, et al. Rhizobial biogeography and inoculation application to soybean in four regions across China [J]. J Appl Microbiol, 2018, 125(3): 853-866. |
| [5] | Tian CF, Zhou YJ, Zhang YM, et al. Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations [J]. Proc Natl Acad Sci USA, 2012, 109(22): 8629-8634. |
| [6] | Balhana RJC, Singla A, Sikder MH, et al. Global analyses of TetR family transcriptional regulators in mycobacteria indicates conservation across species and diversity in regulated functions [J]. BMC Genomics, 2015, 16(1): 479. |
| [7] | Ramos JL, Martínez-Bueno M, Molina-Henares AJ, et al. The TetR family of transcriptional repressors [J]. Microbiol Mol Biol Rev, 2005, 69(2): 326-356. |
| [8] | 吴攀攀, 李博文, 陈克涛, 等. TetR家族转录调控因子配体的研究进展 [J]. 生物工程学报, 2021, 37(7): 2379-2392. |
| Wu PP, Li BW, Chen KT, et al. Ligands of TetR family transcriptional regulators: a review [J]. Chin J Biotechnol, 2021, 37(7): 2379-2392. | |
| [9] | Patil RS, Sharma S, Bhaskarwar AV, et al. TetR and OmpR family regulators in natural product biosynthesis and resistance [J]. Proteins, 2025, 93(1): 38-71. |
| [10] | Ohkama-Ohtsu N, Honma H, Nakagome M, et al. Growth Rate of and Gene Expression in Bradyrhizobium diazoefficiens USDA110 due to a Mutation in blr7984, a TetR Family Transcriptional Regulator Gene [J]. Microbes Environ, 2016, 31(3): 249-259. |
| [11] | Han F, He XQ, Chen WW, et al. Involvement of a novel TetR-like regulator (BdtR) of Bradyrhizobium diazoefficiens in the efflux of isoflavonoid genistein [J]. Mol Plant Microbe Interact, 2020, 33(12): 1411-1423. |
| [12] | Luo RB, Liu BH, Xie YL, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler [J]. Gigascience, 2012, 1(1): 18. |
| [13] | Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics [J]. Genome Res, 2009, 19(9): 1639-1645. |
| [14] | Lombard V, Golaconda Ramulu H, Drula E, et al. The carbohydrate-active enzymes database (CAZy) in 2013 [J]. Nucleic Acids Res, 2014, 42(Database issue): D490-D495. |
| [15] | Zhang XX, Guo HJ, Jiao J, et al. Pyrosequencing of rpoB uncovers a significant biogeographical pattern of rhizobial species in soybean rhizosphere [J]. J Biogeogr, 2017, 44(7): 1491-1499. |
| [16] | 张星星. 大豆慢生根瘤菌分子进化学分析及土著大豆根瘤菌生物地理分布的高通量研究 [D]. 北京: 中国农业大学, 2014. |
| Zhang XX. Molecular evolution of soybean nodulating Bradyrhizobium and biogeography of indigenous soybean rhizobia revealed by amplicon pyrosequencing [D]. Beijing: China Agricultural University, 2014. | |
| [17] | Wang CH, Li M, Zhao Y, et al. SHORT-ROOT paralogs mediate feedforward regulation of D-type cyclin to promote nodule formation in soybean [J]. Proc Natl Acad Sci USA, 2022, 119(3): e2108641119. |
| [18] | Bender FR, Nagamatsu ST, Delamuta JRM, et al. Genetic variation in symbiotic islands of natural variant strains of soybean Bradyrhizobium japonicum and Bradyrhizobium diazoefficiens differing in competitiveness and in the efficiency of nitrogen fixation [J]. Microb Genom, 2022, 8(4): 000795. |
| [19] | Colclough AL, Scadden J, Blair JA. TetR-family transcription factors in Gram-negative bacteria: conservation, variation and implications for efflux-mediated antimicrobial resistance [J]. BMC Genomics, 2019, 20(1): 731. |
| [20] | Singh P, Jain A, Chhabra R, et al. TetR family transcriptional regulators: Lipid metabolism and drug resistance in mycobacteria [J]. Gene Rep, 2024, 36: 101938. |
| [21] | Henríquez T, Stein NV, Jung H. Resistance to bipyridyls mediated by the TtgABC efflux system in Pseudomonas putida KT2440 [J]. Front Microbiol, 2020, 11: 1974. |
| [22] | Watanabe S, Shimada N, Tajima K, et al. Identification and characterization of L-arabonate dehydratase, L-2-keto-3-deoxyarabonate dehydratase, and L-arabinolactonase involved in an alternative pathway of L-arabinose metabolism. Novel evolutionary insight into sugar metabolism [J]. J Biol Chem, 2006, 281(44): 33521-33536. |
| [23] | Nguyen Le Minh P, de Cima S, Bervoets I, et al. Ligand binding specificity of RutR, a member of the TetR family of transcription regulators in Escherichia coli [J]. FEBS Open Bio, 2015, 5: 76-84. |
| [24] | Kendall SL, Burgess P, Balhana R, et al. Cholesterol utilization in mycobacteria is controlled by two TetR-type transcriptional regulators: kstR and kstR2 [J]. Microbiology, 2010, 156(Pt 5): 1362-1371. |
| [25] | Erni B, Siebold C, Christen S, et al. Small substrate, big surprise: fold, function and phylogeny of dihydroxyacetone kinases [J]. Cell Mol Life Sci, 2006, 63(7/8): 890-900. |
| [26] | Li YJ, Wang CH, Zheng L, et al. Natural variation of GmRj2/Rfg1 determines symbiont differentiation in soybean [J]. Curr Biol, 2023, 33(12): 2478-2490.e5. |
| [27] | Wani SR, Dubey AA, Jain V. Ms6244 is a novel Mycobacterium smegmatis TetR family transcriptional repressor that regulates cell growth and morphophysiology [J]. FEBS Lett, 2023, 597(10): 1428-1440. |
| [28] | Lei YK, Asamizu S, Ishizuka T, et al. Regulation of multidrug efflux pumps by TetR family transcriptional repressor negatively affects secondary metabolism in Streptomyces coelicolor A3(2) [J]. Appl Environ Microbiol, 2023, 89(3): e0182222. |
| [29] | Schmacht M, Lorenz E, Senz M. Microbial production of glutathione [J]. World J Microbiol Biotechnol, 2017, 33(6): 106. |
| [30] | Luo S, Yin J, Peng Y, et al. Glutathione is involved in detoxification of peroxide and root nodule symbiosis of Mesorhizobium huakuii [J]. Curr Microbiol, 2020, 77(1): 1-10. |
| [31] | Van Laar TA, Esani S, Birges TJ, et al. Pseudomonas aeruginosa gshA mutant is defective in biofilm formation, swarming, and pyocyanin production [J]. mSphere, 2018, 3(2): e00155-18. |
| [32] | Ku JWK, Gan YH. New roles for glutathione: Modulators of bacterial virulence and pathogenesis [J]. Redox Biol, 2021, 44: 102012. |
| [1] | 吕镇, 甘恬, 霍思羽, 赵晨笛, 赵梦瑶, 李亚涛, 马玉超, 耿玉清. 产Surfactin贝莱斯芽胞杆菌C5A-1的鉴定和所产Surfactin对植物的促生效果[J]. 生物技术通报, 2025, 41(9): 265-276. |
| [2] | 翟莹, 计俊杰, 陈炯辛, 于海伟, 李珊珊, 赵艳, 马天意. 异源过表达大豆GmNF-YB24提高转基因烟草抗旱性[J]. 生物技术通报, 2025, 41(8): 137-145. |
| [3] | 朱丽娟, 张锴, 温晓蕾, 褚佳豪, 史凤玉, 王艳丽. 基于WGCNA挖掘野生大豆耐镉关键基因[J]. 生物技术通报, 2025, 41(8): 124-136. |
| [4] | 牛景萍, 赵婧, 郭茜, 王书宏, 赵晋忠, 杜维俊, 殷丛丛, 岳爱琴. 基于WGCNA鉴定大豆抗大豆花叶病毒NAC转录因子及其诱导表达分析[J]. 生物技术通报, 2025, 41(7): 95-105. |
| [5] | 赵强, 陈思宇, 彭方丽, 汪灿, 高杰, 周棱波, 张国兵, 姜昱雯, 邵明波. 间作与施氮对高粱根际土壤细菌多样性及功能的影响[J]. 生物技术通报, 2025, 41(6): 307-316. |
| [6] | 谭玉荣, 陈东亮, 杨守臻, 赖振光, 唐向民, 孙祖东, 曾维英. 大豆抗豆卷叶螟GmKTI1-like的功能研究[J]. 生物技术通报, 2025, 41(6): 99-108. |
| [7] | 张慧, 卢文才, 王冬, 刘倩, 马连杰. 一株高产吲哚乙酸的Bacillus cereus YT2-1C的鉴定及促生作用[J]. 生物技术通报, 2025, 41(5): 300-309. |
| [8] | 吴泽银, 黄晨阳, 赵梦然, 张利姣, 姚方杰. 短柄白黄侧耳CCMSSC 04611基因组特异性分析[J]. 生物技术通报, 2025, 41(5): 320-332. |
| [9] | 陈永旗, 李志文, 李鑫, 原若曦, 王春旭, 韩毅强, 高亚梅. 黑土区大豆根际土壤放线菌的分离与功能研究[J]. 生物技术通报, 2025, 41(5): 255-266. |
| [10] | 李志强, 罗正乾, 徐琳黎, 周国慧, 屈丝雨, 刘恩良, 顼东婷. 基于T2T基因组鉴定大豆R2R3-MYB基因家族及干旱和盐胁迫下的表达分析[J]. 生物技术通报, 2025, 41(5): 141-152. |
| [11] | 赵婧, 郭茜, 李睿琦, 雷滢炀, 岳爱琴, 赵晋忠, 殷丛丛, 杜维俊, 牛景萍. 大豆GmGST基因簇基因序列分析及诱导表达分析[J]. 生物技术通报, 2025, 41(5): 129-140. |
| [12] | 林紫依, 吴一舟, 叶芳贤, 朱淑颖, 刘燕敏, 刘骕骦. 大豆GmPM31基因启动子响应高温高湿胁迫的功能分析[J]. 生物技术通报, 2025, 41(3): 90-97. |
| [13] | 宋英培, 王灿, 周会汶, 孔可可, 许孟歌, 王瑞凯. 基于全基因组关联分析和遗传多样性的大豆裂荚性状解析[J]. 生物技术通报, 2025, 41(2): 97-106. |
| [14] | 刘克寒, 杨升辉, 黄巧云, 崔文靖. 黑龙江大豆根瘤菌及根际促共生菌株的筛选及应用[J]. 生物技术通报, 2025, 41(1): 252-262. |
| [15] | 张婷, 万雨欣, 徐伟慧, 王志刚, 陈文晶, 胡云龙. 一株玉米根际促生菌Leclercia adecarboxylata LN01促生效果研究及其基因组分析[J]. 生物技术通报, 2025, 41(1): 263-275. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||