








生物技术通报 ›› 2025, Vol. 41 ›› Issue (9): 44-53.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0222
王芳( ), 邵会茹, 吕林龙, 赵点, 胡振, 吕建珍(
), 邵会茹, 吕林龙, 赵点, 胡振, 吕建珍( ), 姜亮(
), 姜亮( )
)
收稿日期:2025-03-04
出版日期:2025-09-26
发布日期:2025-09-24
通讯作者:
吕建珍,女,硕士,研究员,研究方向 :谷子产量和品质遗传改良;E-mail: lvjianzhen110@163.com作者简介:王芳,女,硕士,研究方向 :谷子高产基因鉴定;E-mail: 2897447253@qq.com
基金资助:
WANG Fang( ), SHAO Hui-ru, LYU Lin-long, ZHAO Dian, HU Zhen, LYU Jian-zhen(
), SHAO Hui-ru, LYU Lin-long, ZHAO Dian, HU Zhen, LYU Jian-zhen( ), JIANG Liang(
), JIANG Liang( )
)
Received:2025-03-04
Published:2025-09-26
Online:2025-09-24
摘要:
目的 TurboID邻近蛋白标记技术是一种基于生物素连接酶的新型蛋白互作研究技术,具有标记速度快、时空分辨率高和毒性小等优点。目前该技术仍处于逐步推广阶段,并在少数物种中得到应用。通过构建多种代表性物种的TurboID表达系统,评估其在单子叶植物、双子叶植物及原核生物中的标记效能和应用范围。 方法 分别构建在单子叶植物[水稻(Oryza sativa)]、双子叶植物[拟南芥(Arabidopsis thaliana)、番茄(Solanum lycopersicum)、本氏烟草(Nicotiana benthamiana)]和细菌[大肠杆菌(Escherichia coli)]中表达“标签蛋白+TurboID”融合蛋白的载体,并将其转化至相应物种。通过生物素溶液处理转基因材料后提取总蛋白,利用标签蛋白免疫印迹检测融合蛋白的表达情况,并通过生物素免疫印迹检测内源蛋白的标记情况。 结果 在植物中采用泛素启动子、细菌中采用T7启动子驱动表达,实现了“标签蛋白+TurboID”融合蛋白在拟南芥、番茄、烟草、水稻愈伤组织和大肠杆菌中的高效表达。免疫印迹显示融合蛋白表达良好,且经生物素处理后,各系统中均有多种内源蛋白被有效标记,表明所构建的TurboID系统在单子叶植物、双子叶植物及原核生物中均具有良好适用性和推广潜力。 结论 成功在单子叶植物、双子叶植物和细菌中建立了TurboID邻近蛋白标记方法,为复杂蛋白互作网络分析提供了高效、可靠的技术平台。
王芳, 邵会茹, 吕林龙, 赵点, 胡振, 吕建珍, 姜亮. 植物和细菌TurboID邻近蛋白标记方法的建立[J]. 生物技术通报, 2025, 41(9): 44-53.
WANG Fang, SHAO Hui-ru, LYU Lin-long, ZHAO Dian, HU Zhen, LYU Jian-zhen, JIANG Liang. Establishment of TurboID Proximity Labeling Technology in Plants and Bacteria[J]. Biotechnology Bulletin, 2025, 41(9): 44-53.
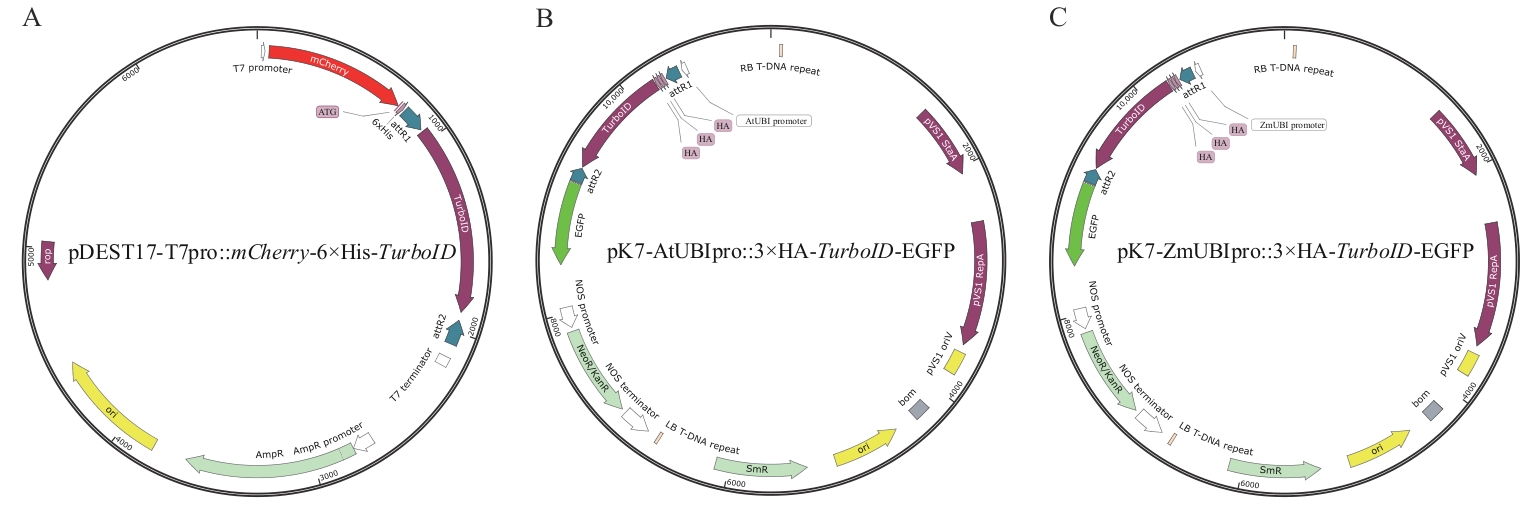
图1 植物和细菌TurboID邻近蛋白标记载体A:大肠杆菌TurboID邻近蛋白标记载体pDEST17-T7pro::mCherry-6×His-TurboID;B:双子叶植物(拟南芥、番茄和烟草)TurboID邻近蛋白标记载体pK7-AtUBIpro::3×HA-TurboID-EGFP;C:单子叶植物(水稻)TurboID邻近蛋白标记载体pK7-ZmUBIpro::3×HA-TurboID-EGFP
Fig. 1 Expression vectors for TurboID proximity labeling in plants and bacteriaA: Expression vector (pDEST17-T7pro::mCherry-6×His-TurboID) for TurboID proximity labeling in Escherichia coli. B: Expression vector (pK7-AtUBIpro::3×HA-TurboID-EGFP) for TurboID proximity labeling in dicotyledonous plants, including Arabidopsis thalian, Solanum lycopersicum, and Nicotiana benthamiana. C: Expression vector (pK7-ZmUBIpro::3×HA-TurboID-EGFP) for TurboID proximity labeling in monocotyledonous plant (Oryza sativa)
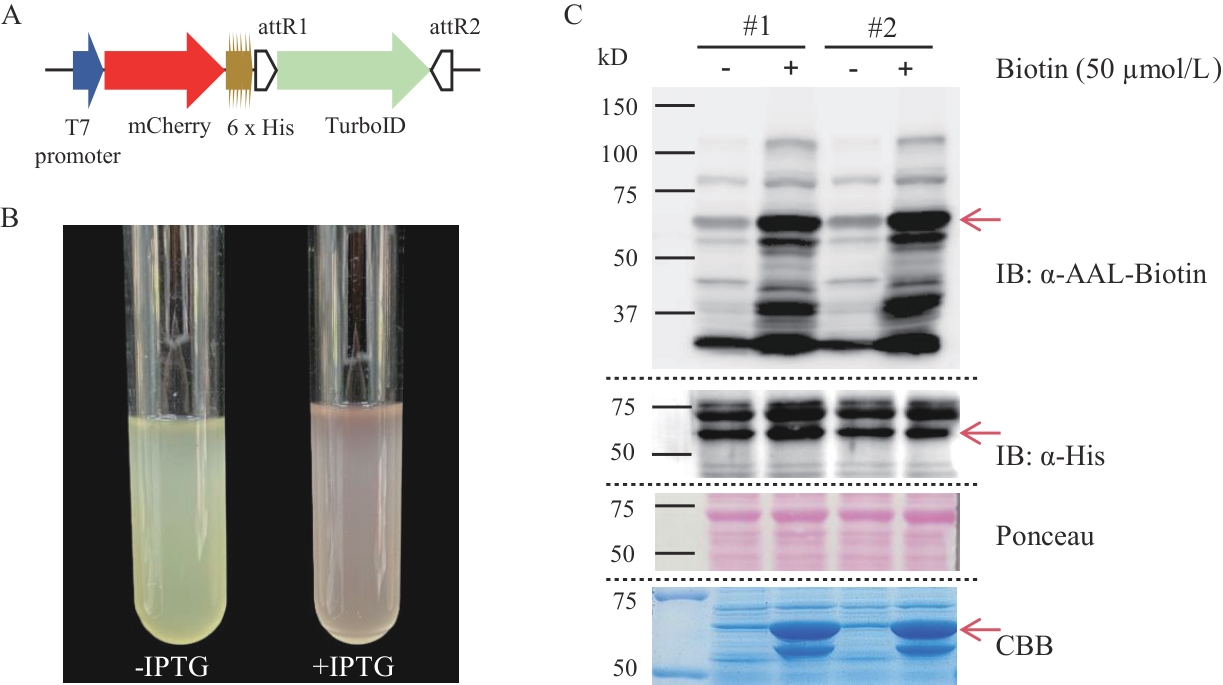
图3 大肠杆菌TurboID邻近蛋白标记方法的创建A:融合蛋白mCherry-TurboID的表达元件示意图;B:添加和不添加IPTG诱导剂的菌液照片;C:蛋白免疫印迹检测2个转基因大肠杆菌菌种(#1; #2)中融合蛋白的表达和邻近蛋白的生物素标记。“-”和“+”分别表示用ddH2O和50 μmol/L生物素溶液处理,在考马斯亮蓝染色图中分别表示未诱导和诱导后
Fig. 3 Establishment of the TurboID proximity labeling method in E. coliA: Schematic diagram of the expression elements of the fusion protein mCherry-TurboID3. B: Photographs of bacterial cultures with and without IPTG induction. C: Western-blot detection of the expression of the fusion protein and biotinylation of proximity labeling in two transgenic E. coli strains (#1; #2). "-" and "+" indicate treatment with ddH2O and 50 μmol/L biotin solution respectively, in the Coomassie Brilliant Blue staining image, they indicate the uninduced and induced states respectively

图4 邻近蛋白标记中生物素浓度与处理时间的优化A:使用不同浓度的生物素溶液对诱导后菌液处理3 h,使用α-AAL-Biotin和α-His抗体进行蛋白免疫印迹检测;B:使用50 μmol/L生物素溶液对诱导后菌液处理不同时间,进行蛋白免疫印迹检测
Fig. 4 Optimization of biotin concentration and labeling time used in proximity labelingA: Treat the induced bacterial culture with biotin solutions of different concentrations for 3 h and then perform Western-blot detection using α-AAL-Biotin and α-His antibodies. B: Treat the induced bacterial culture with a 50 μmol/L biotin solution for different durations and then perform Western-blot detection

图5 拟南芥TurboID邻近蛋白标记方法的创建A:融合蛋白TurboID-EGFP的表达元件示意图;B:野生型拟南芥与转基因拟南芥30 d幼苗图;C:蛋白免疫印迹检测2个转基因拟南芥株系(#3; #5)中融合蛋白的表达和蛋白的生物素标记。“-”和“+”分别表示用ddH2O和100 μmol/L生物素溶液处理。下同
Fig. 5 Establishment of the TurboID proximity labeling method in ArabidopsisA: Schematic diagram of the expression elements of the fusion protein TurboID-EGFP. B: Phenotypic comparison of 30-day-old seedlings between wild-type and transgenic Arabidopsis thaliana. C: Western-blot detection of the expression of fusion proteins and biotinylation of proximity labeling in two transgenic Arabidopsis lines (#3; #5). "-" and "+" indicate treatment with ddH2O and 100 μmol/L biotin solution respectively. The same below

图6 番茄TurboID邻近蛋白标记方法的创建A:野生型番茄与转基因番茄30 d幼苗图;B:蛋白免疫印迹检测2个转基因番茄株系(#1; #2)叶片融合蛋白的表达和邻近蛋白的生物素标记
Fig. 6 Establishment of the TurboID proximity labeling method in S. lycopersicumA: Comparative analysis of 30-day-old wild-type and transgenic Solanum lycopersicum. B: Western-blot detection of the expression of fusion proteins and biotinylation of proximity labeling in the leaves of two transgenic tomato lines (#1; #2)

图7 基于烟草瞬时表达的TurboID邻近蛋白标记方法的创建A:烟草叶片注射图片,Ruby表达农杆菌注射烟草作为烟草表达情况的可视化对照,CK为未注射农杆菌的叶片;B:蛋白免疫印迹检测融合蛋白的表达和邻近蛋白的生物素标记;从左到右3个样品分别是注射农杆菌但不注射生物素的叶片、未注射任何农杆菌的叶片和注射农杆菌且注射生物素(20 μmol/L)的烟草叶片
Fig.7 Establishment of the TurboID proximity labeling method based on tobacco transient expressionA: Image of tobacco leaf injection. Ruby-expressing Agrobacterium-injected tobacco serves as a visual control for tobacco expression, and CK being the leaf not injected with any Agrobacterium. B: Western-blot detection of the expression of fusion proteins and biotinylation of proximity labeling. From left to right, the three samples are tobacco leaves injected with Agrobacterium but not biotin, leaves not injected with any Agrobacterium or biotin, and tobacco leaves injected with both Agrobacterium and 20 μmol/L biotin
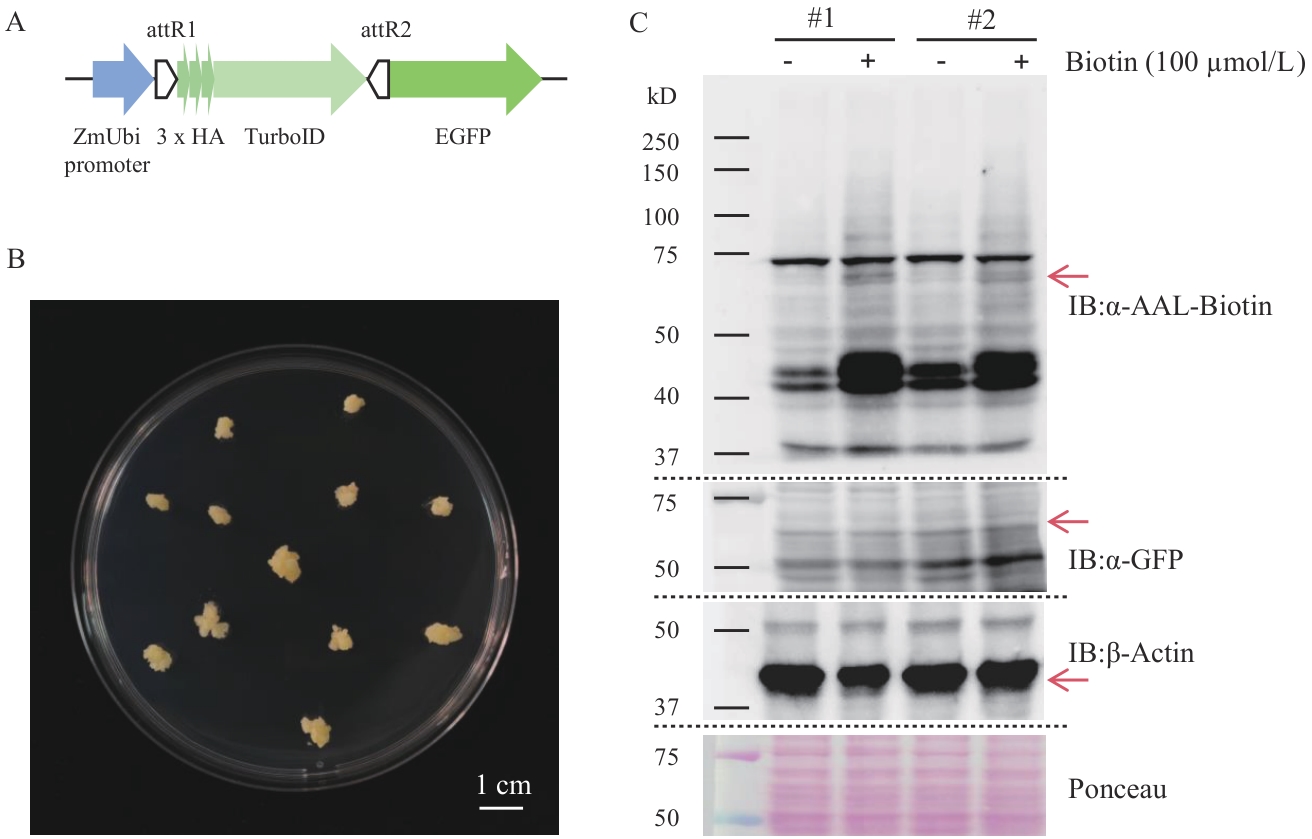
图8 水稻愈伤TurboID邻近蛋白标记方法的创建A:融合蛋白TurboID-EGFP的表达元件示意图;B:水稻愈伤图;C:蛋白免疫印迹检测2个转基因水稻株系(#1; #2)中融合蛋白的表达和邻近蛋白的生物素标记
Fig. 8 Establishment of the TurboID proximity labeling method in rice callusesA: Schematic diagram of the expression elements of the fusion protein TurboID-EGFP. B: Image of rice calluses. C: Western-blot detection of the expression of the fusion protein and biotinylation of proximity labeling in two transgenic rice lines (#1; #2)
| [1] | Gingras AC, Abe KT, Raught B. Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles [J]. Curr Opin Chem Biol, 2019, 48: 44-54. |
| [2] | Duarte CEM, Euclydes NC. Protein-protein interaction via two-hybrid assay in yeast [J]. Methods Mol Biol, 2024, 2724: 193-210. |
| [3] | Lo SF. Co-immunoprecipitation (co-ip) in mammalian cells [J]. Methods Mol Biol, 2023, 2655: 67-77. |
| [4] | Zhang YJ, Natale R, Domingues Júnior AP, et al. Rapid identification of protein-protein interactions in plants [J]. Curr Protoc Plant Biol, 2019, 4(4): e20099. |
| [5] | Douzi B. Protein-protein interactions: surface plasmon resonance [J]. Methods Mol Biol, 2017, 1615: 257-275. |
| [6] | Roux KJ, Kim DI, Raida M, et al. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells [J]. J Cell Biol, 2012, 196(6): 801-810. |
| [7] | Choi CR, Rhee HW. Proximity labeling: an enzymatic tool for spatial biology [J]. Trends Biotechnol, 2022, 40(2): 145-148. |
| [8] | Sato S, Yoshida M, Hatano K, et al. N′-acyl-N-methylphenylenediamine as a novel proximity labeling agent for signal amplification in immunohistochemistry [J]. Bioorg Med Chem, 2019, 27(6): 1110-1118. |
| [9] | Rhee HW, Zou P, Udeshi ND, et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging [J]. Science, 2013, 339(6125): 1328-1331. |
| [10] | Loh KH, Stawski PS, Draycott AS, et al. Proteomic analysis of unbounded cellular compartments: synaptic clefts [J]. Cell, 2016, 166(5): 1295-1307.e21. |
| [11] | Hopkins C, Gibson A, Stinchcombe J, et al. Chimeric molecules employing horseradish peroxidase as reporter enzyme for protein localization in the electron microscope [J]. Meth Enzymol, 2000, 327: 35-45. |
| [12] | Martell JD, Deerinck TJ, Sancak Y, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy [J]. Nat Biotechnol, 2012, 30(11): 1143-1148. |
| [13] | Trinkle-Mulcahy L. Recent advances in proximity-based labeling methods for interactome mapping [J]. F1000Res, 2019, 8(F1000 Faculty Rev): 135. |
| [14] | Parrott MB, Barry MA. Metabolic biotinylation of secreted and cell surface proteins from mammalian cells [J]. Biochem Biophys Res Commun, 2001, 281(4): 993-1000. |
| [15] | Jeong KH, Son SB, Ko JH, et al. Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor [J]. Biochem Biophys Res Commun, 2024, 733: 150601. |
| [16] | Kim DI, Roux KJ. Filling the void: proximity-based labeling of proteins in living cells [J]. Trends Cell Biol, 2016, 26(11): 804-817. |
| [17] | Kim DI, Jensen SC, Noble KA, et al. An improved smaller biotin ligase for BioID proximity labeling [J]. Mol Biol Cell, 2016, 27(8): 1188-1196. |
| [18] | Branon TC, Bosch JA, Sanchez AD, et al. Efficient proximity labeling in living cells and organisms with TurboID [J]. Nat Biotechnol, 2018, 36(9): 880-887. |
| [19] | Zhang YL, Song GY, Lal NK, et al. TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity [J]. Nat Commun, 2019, 10(1): 3252. |
| [20] | Mair A, Xu SL, Branon TC, et al. Proximity labeling of protein complexes and cell-type-specific organellar proteomes in Arabidopsis enabled by TurboID [J]. eLife, 2019, 8: e47864. |
| [21] | Arora D, Abel NB, Liu C, et al. Establishment of proximity-dependent biotinylation approaches in different plant model systems [J]. Plant Cell, 2020, 32(11): 3388-3407. |
| [22] | Kreis E, König K, Misir M, et al. TurboID reveals the proxiomes of Chlamydomonas proteins involved in thylakoid biogenesis and stress response [J]. Plant Physiol, 2023, 193(3): 1772-1796. |
| [23] | Sun FA, Hamada N, Montes C, et al. TurboID-based proteomic profiling reveals proxitome of ASK1 and CUL1 of the SCF ubiquitin ligase in plants [J]. New Phytol, 2024, 244(6): 2127-2136. |
| [24] | Li PP, Li JJ, Wang L, et al. Proximity labeling of interacting proteins: application of BioID as a discovery tool [J]. Proteomics, 2017, 17(20): 1700002. |
| [25] | Cho KF, Branon TC, Udeshi ND, et al. Proximity labeling in mammalian cells with TurboID and split-TurboID [J]. Nat Protoc, 2020, 15(12): 3971-3999. |
| [26] | 邝嘉怡, 李洪清, 沈文锦, 等. 基于TurboID的植物蛋白邻近标记实验方法 [J]. 植物学报, 2021, 56(5): 584-593. |
| Kuang JY, Li HQ, Shen WJ, et al. Methods for TurboID-based proximal labeling in plants [J]. Chin Bull Bot, 2021, 56(5): 584-593. | |
| [27] | 刘佳欣, 何明良, 刘颖湘, 等. 利用TurboID邻近蛋白标记技术获得水稻互作蛋白组的实验方法 [J]. 土壤与作物, 2023, 12(3): 256-263. |
| Liu JX, He ML, Liu YX, et al. Experimental method for obtaining interaction proteome using TurboID-based proximity labeling technology in rice [J]. Soils Crops, 2023, 12(3): 256-263. | |
| [28] | May DG, Scott KL, Campos AR, et al. Comparative application of BioID and TurboID for protein-proximity biotinylation [J]. Cells, 2020, 9(5): 1070. |
| [29] | Tan H, Zhou Y, Dinius E, et al. The Ti-TAN plasmid toolbox for TurboID-based proximity labeling assays in Nicotiana benthamiana [J]. J Integr Plant Biol, 2024, 66(2): 166-168. |
| [30] | Cho KF, Branon TC, Rajeev S, et al. Split-TurboID enables contact-dependent proximity labeling in cells [J]. Proc Natl Acad Sci USA, 2020, 117(22): 12143-12154. |
| [31] | Strotmann VI, Stahl Y. Visualization of in vivo protein-protein interactions in plants [J]. J Exp Bot, 2022, 73(12): 3866-3880. |
| [32] | Garloff V, Krüger T, Brakhage A, et al. Control of TurboID-dependent biotinylation intensity in proximity ligation screens [J]. J Proteom, 2023, 279: 104886. |
| [33] | Yang XX, Wen ZY, Zhang DL, et al. Proximity labeling: an emerging tool for probing in planta molecular interactions [J]. Plant Commun, 2020, 2(2): 100137. |
| [34] | Zhang YL, Li YY, Yang XX, et al. TurboID-based proximity labeling for in planta identification of protein-protein interaction networks [J]. J Vis Exp, 2020(159): 10.3791/60728. |
| [35] | Feng C, Roitinger E, Hudecz O, et al. TurboID-based proteomic profiling of meiotic chromosome axes in Arabidopsis thaliana [J]. Nat Plants, 2023, 9(4): 616-630. |
| [36] | Li XF, Wei YP, Fei QL, et al. TurboID-mediated proximity labeling for screening interacting proteins of FIP37 in Arabidopsis [J]. Plant Direct, 2023, 7(12): e555. |
| [37] | Xiong ZR, Lo HP, McMahon KA, et al. In vivo proteomic mapping through GFP-directed proximity-dependent biotin labelling in zebrafish [J]. eLife, 2021, 10: e64631. |
| [38] | Kim HB, Kim KE. Precision proteomics with TurboID: mapping the suborganelle landscape [J]. Korean J Physiol Pharmacol, 2024, 28(6): 495-501. |
| [39] | Merta H, Gov K, Isogai T, et al. Spatial proteomics of ER tubules reveals CLMN, an ER-actin tether at focal adhesions that promotes cell migration [J]. Cell Rep, 2025, 44(4): 115502. |
| [1] | 李雅琼, 格桑拉毛, 陈启迪, 杨宇环, 何花转, 赵耀飞. 异源过表达高粱SbSnRK2.1增强拟南芥对盐胁迫的抗性[J]. 生物技术通报, 2025, 41(8): 115-123. |
| [2] | 康琴, 汪霞, 谌明洋, 徐静天, 陈诗兰, 廖平杨, 许文志, 吴卫, 徐东北. 薄荷UV-B受体基因McUVR8的克隆与表达分析[J]. 生物技术通报, 2025, 41(8): 255-266. |
| [3] | 邓美壁, 严浪, 詹志田, 朱敏, 和玉兵. RUBY辅助的水稻高效CRISPR基因编辑[J]. 生物技术通报, 2025, 41(8): 65-73. |
| [4] | 侯鹰翔, 费思恬, 黎妮, 李兰, 宋松泉, 王伟平, 张超. 水稻miRNAs响应生物胁迫研究进展[J]. 生物技术通报, 2025, 41(7): 69-80. |
| [5] | 王从欢, 伍国强, 魏明. 植物CBL调控逆境胁迫响应的作用机制[J]. 生物技术通报, 2025, 41(7): 1-16. |
| [6] | 黄丹丹, 吴云翼, 邹建华, 俞婷, 朱炎辉, 杨梅宏, 董文丽, 高冬丽. 马铃薯StPTST2a基因的克隆及互作分析[J]. 生物技术通报, 2025, 41(7): 172-180. |
| [7] | 吴浩, 董伟峰, 贺子天, 李艳肖, 谢辉, 孙明哲, 沈阳, 孙晓丽. 水稻BXL基因家族的全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(6): 87-98. |
| [8] | 杜量衡, 唐黄磊, 张治国. 控制水稻光响应基因ELM1的图位克隆[J]. 生物技术通报, 2025, 41(5): 82-89. |
| [9] | 刘园园, 陈析丰, 钱前, 高振宇. 水稻穗发育调控的分子机制研究进展[J]. 生物技术通报, 2025, 41(5): 1-13. |
| [10] | 陈晓军, 惠建, 马洪文, 白海波, 钟楠, 李稼润, 樊云芳. 利用单碱基基因编辑技术创制OsALS抗除草剂水稻种质资源[J]. 生物技术通报, 2025, 41(4): 106-114. |
| [11] | 刘彤彤, 李肖慧, 杨骏龙, 陈旺, 玉猛, 王超凡, 王凤茹, 客绍英. ZmSTART1调控玉米维管束建成的功能研究[J]. 生物技术通报, 2025, 41(4): 115-122. |
| [12] | 李欣芃, 张武汉, 张莉, 舒服, 何强, 郭杨, 邓华凤, 王悦, 孙平勇. γ射线诱变创制水稻突变体及其分子鉴定[J]. 生物技术通报, 2025, 41(3): 35-43. |
| [13] | 方慧敏, 顾艺枢, 张晶, 张龙. 水稻叶片淀粉的分离与理化性质分析[J]. 生物技术通报, 2025, 41(2): 51-57. |
| [14] | 田栩瑞, 霍信屹, 郭云涵, 向林, 产祝龙, 王艳平. 百合LoSAUR10基因的表达特征及功能分析[J]. 生物技术通报, 2025, 41(1): 221-229. |
| [15] | 金素奎, 国倩倩, 刘巧泉, 高继平. 一种水稻叶片基因组DNA简易提取方法[J]. 生物技术通报, 2025, 41(1): 74-84. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||