








生物技术通报 ›› 2025, Vol. 41 ›› Issue (4): 115-122.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0981
• 研究报告 • 上一篇
刘彤彤( ), 李肖慧, 杨骏龙, 陈旺, 玉猛, 王超凡, 王凤茹(
), 李肖慧, 杨骏龙, 陈旺, 玉猛, 王超凡, 王凤茹( ), 客绍英(
), 客绍英( )
)
收稿日期:2024-10-08
出版日期:2025-04-26
发布日期:2025-04-25
通讯作者:
王凤茹,女,博士,教授,研究方向 :植物生长发育调控;E-mail: wfr15931945160@126.com作者简介:刘彤彤,女,硕士,研究方向 :植物生长发育调控;E-mail: 873742820@qq.com
基金资助:
LIU Tong-tong( ), LI Xiao-hui, YANG Jun-long, CHEN Wang, YU Meng, WANG Chao-fan, WANG Feng-ru(
), LI Xiao-hui, YANG Jun-long, CHEN Wang, YU Meng, WANG Chao-fan, WANG Feng-ru( ), KE Shao-ying(
), KE Shao-ying( )
)
Received:2024-10-08
Published:2025-04-26
Online:2025-04-25
摘要:
目的 解析玉米基因ZmSTART1的结构,明确ZmSTART1的表达特性,分析ZmSTART1在维管束建成过程中的作用,为玉米抗倒伏和产量性状的遗传改良提供理论基础。 方法 利用生物信息学方法,解析ZmSTART1的结构特征;利用Real-Time PCR技术分析ZmSTART1的时空表达特性;利用ZmSTART1-GFP融合技术转化烟草叶片,对ZmSTART1进行亚细胞定位;构建ZmSTART1过表达载体,利用农杆菌侵染花絮法转化Col-0拟南芥,利用Real-Time PCR技术验证过表达ZmSTART1拟南芥阳性苗,观察过表达ZmSTART1拟南芥的维管束特征,明确ZmSTART1调控维管束建成的生物学功能。 结果 生物信息学分析发现,ZmSTART1是一个仅含有1个START结构域的疏水性蛋白质。Real-Time PCR技术分析ZmSTART1的时空表达特性发现,ZmSTART1在花药、叶片和第一节间中表达量较高。亚细胞定位表明,ZmSTART1定位于细胞质膜上。创制过表达ZmSTART1的拟南芥植株ZmSTART1-OE发现,过表达ZmSTART1-OE转基因拟南芥子叶叶脉和真叶二、三级叶脉个数均比野生型拟南芥多,而且叶脉形成的封闭空间数也显著高于野生型对照,ZmSTART1-OE转基因拟南芥叶片出现四级叶脉时,野生型对照叶片只有三级叶脉。观察茎维管束发育情况发现,野生型拟南芥茎初生结构内维管束为6个,ZmSTART1-OE茎初生结构维管束增多为7个,束间纤维细胞层数减少、番红染色变浅、木质素含量降低。 结论 ZmSTART1属于START家族成员,定位于细胞质膜,在玉米的维管束建成过程中具有重要作用。
刘彤彤, 李肖慧, 杨骏龙, 陈旺, 玉猛, 王超凡, 王凤茹, 客绍英. ZmSTART1调控玉米维管束建成的功能研究[J]. 生物技术通报, 2025, 41(4): 115-122.
LIU Tong-tong, LI Xiao-hui, YANG Jun-long, CHEN Wang, YU Meng, WANG Chao-fan, WANG Feng-ru, KE Shao-ying. Functional Study on ZmSTART1 Regulation of Maize Vascular Bundle Formation[J]. Biotechnology Bulletin, 2025, 41(4): 115-122.
类型 Type | 真叶面积 True leaf area/mm2 | 子叶面积 Cotyledon area/mm2 | 叶柄长 Petiole length/cm | 株高 Plant height/cm | 茎面积 Stem area/mm2 | 茎粗 Stem circumference/mm |
|---|---|---|---|---|---|---|
| WT | 6.15±1.06aa | 8.35±0.64aa | 0.25±0.04aa | 11±1.00aa | 0.62±0.01aa | 2.79±0.03aa |
| ZmSTART1-OE1 | 9.4±0.28AA | 6.15±0.47AA | 0.35±0.05AA | 26.7±1.00AA | 0.67±0.11aa | 2.89±0.05aa |
| ZmSTART1-OE2 | 8.8±0.11AA | 5.87±0.24AA | 0.31±0.08Aa | 22.5±1.87AA | 0.63±0.13aa | 2.81±0.17aa |
| ZmSTART1-OE3 | 8.8±0.23AA | 5.15±0.33AA | 0.28±0.07aa | 18.6±1.65AA | 0.62±0.07aa | 2.79±0.13aa |
表1 形态指标统计学分析
Table 1 Statistical analysis of morphological indexes
类型 Type | 真叶面积 True leaf area/mm2 | 子叶面积 Cotyledon area/mm2 | 叶柄长 Petiole length/cm | 株高 Plant height/cm | 茎面积 Stem area/mm2 | 茎粗 Stem circumference/mm |
|---|---|---|---|---|---|---|
| WT | 6.15±1.06aa | 8.35±0.64aa | 0.25±0.04aa | 11±1.00aa | 0.62±0.01aa | 2.79±0.03aa |
| ZmSTART1-OE1 | 9.4±0.28AA | 6.15±0.47AA | 0.35±0.05AA | 26.7±1.00AA | 0.67±0.11aa | 2.89±0.05aa |
| ZmSTART1-OE2 | 8.8±0.11AA | 5.87±0.24AA | 0.31±0.08Aa | 22.5±1.87AA | 0.63±0.13aa | 2.81±0.17aa |
| ZmSTART1-OE3 | 8.8±0.23AA | 5.15±0.33AA | 0.28±0.07aa | 18.6±1.65AA | 0.62±0.07aa | 2.79±0.13aa |

图1 ZmSTART1结构分析A:ZmSTART1蛋白的结构域示意图;B:ZmSTART1基因的外显子和内含子示意图;C:ZmSTART1的三级结构
Fig. 1 Structure analysis of ZmSTART1A: Schematic diagram of the domain in ZmSTART1 protein. B: Schematic diagram of the exons and introns in ZmSTART1 gene. C: The tertiary structure of ZmSTART1

图2 拟南芥和玉米START家族成员系统发育进化树GRMZM:玉米;At:拟南芥
Fig. 2 Phylogenetic tree analysis of START genes of maize (Zea mays) and A. thalianaGRMZM: Zea mays; At: Arabidopsis thaliana
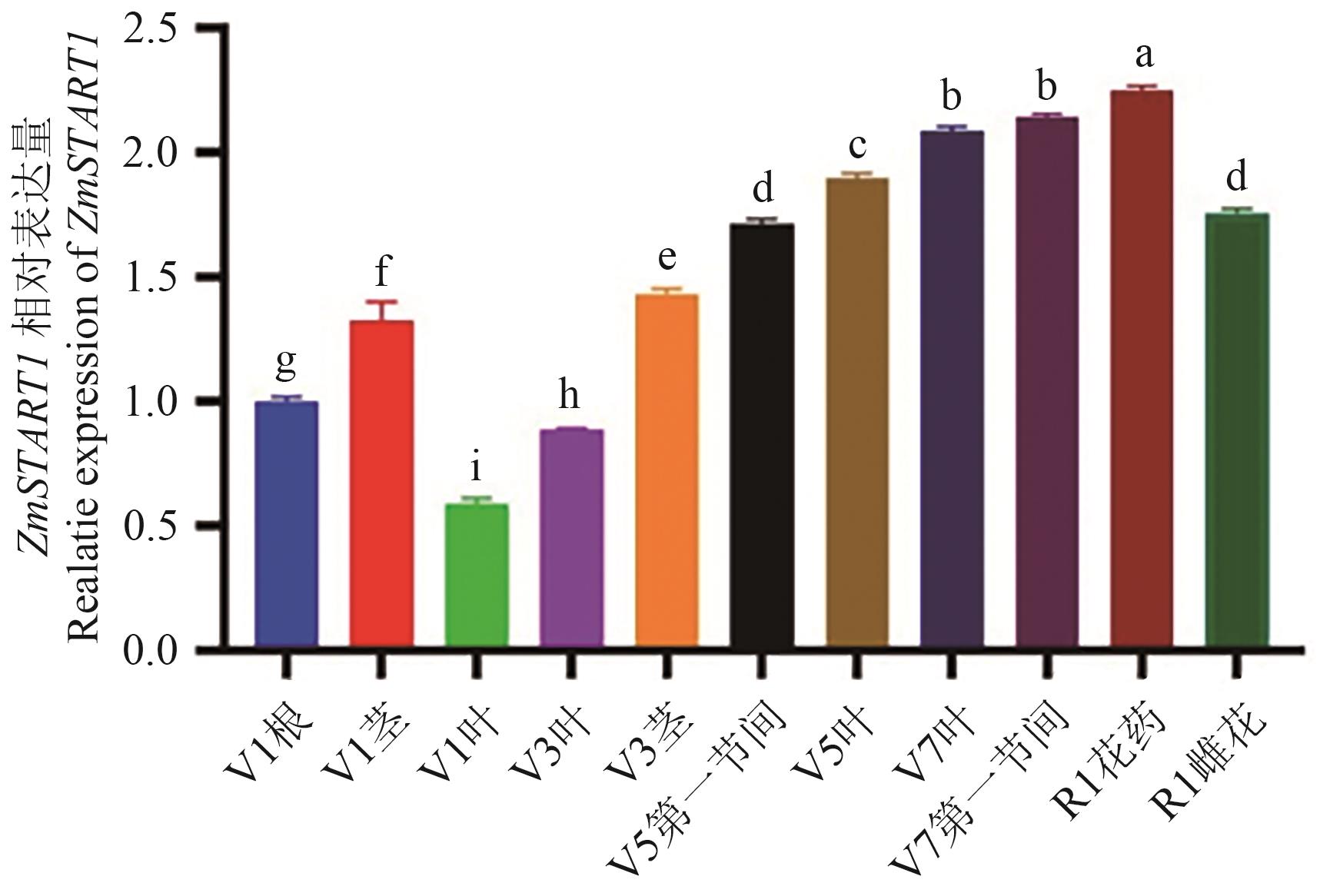
图3 玉米不同组织部位中ZmSTART1的表达V1:1片真叶展开期;V3:3片真叶展开期;V5:5片真叶展开期;V7:7片真叶展开期;R1:玉米的吐丝期。不同小写字母代表差异显著水平≤0.05。下同
Fig. 3 Expressions of ZmSTART1 in different tissues of maizeV1: 1 true leaf development stage. V3: 3 true leaf development stage. V5: 5 true leaf development stage. V7: 7 true leaf development stage. R1: Silking stage of maize. Different lowercase letters indicate a significant difference ≤0.05. The same below

图5 ZmSTART1阳性苗的筛选与鉴定A:潮霉素筛选;B:PCR鉴定;C:RT-qPCR表达量分析
Fig. 5 Screening and identification of ZmSTART1 positive seedlingsA: Hygromycin screening. B: PCR identification. C: Analysis of RT-qPCR expression
| 类型 | 维管束个数 | 子叶二级叶脉 | 真叶二级叶脉数 | 真叶三级叶脉数 | 真叶四级叶脉数 | 子叶脉间区域 |
|---|---|---|---|---|---|---|
| Type | No. of vascular bundles | Cotyledon vein/Veins | Total number of secondary veins in true leaves | Total number of tertiary veins in true leaves | Total number of fourth-order veins of true leaves | Region between veins of cotyledons |
| WT | 6aa | 5±1aa | 12±1aa | 12±1aa | 0aa | 5±1aa |
| ZmSTART1-OE1 | 7±1Aa | 7±1AA | 17±2AA | 37±7AA | 4±1AA | 7±1AA |
| ZmSTART1-OE2 | 7±1Aa | 7±1AA | 16±2AA | 33±8AA | 3±1AA | 6±7Aa |
| ZmSTART1-OE3 | 6±1aa | 6±1Aa | 15±2Aa | 30±8AA | 2±1AA | 5±7aa |
表2 叶片叶脉、茎维管束表型数据统计
Table 2 Phenotype data statistics of superficial veins and vascular bundles
| 类型 | 维管束个数 | 子叶二级叶脉 | 真叶二级叶脉数 | 真叶三级叶脉数 | 真叶四级叶脉数 | 子叶脉间区域 |
|---|---|---|---|---|---|---|
| Type | No. of vascular bundles | Cotyledon vein/Veins | Total number of secondary veins in true leaves | Total number of tertiary veins in true leaves | Total number of fourth-order veins of true leaves | Region between veins of cotyledons |
| WT | 6aa | 5±1aa | 12±1aa | 12±1aa | 0aa | 5±1aa |
| ZmSTART1-OE1 | 7±1Aa | 7±1AA | 17±2AA | 37±7AA | 4±1AA | 7±1AA |
| ZmSTART1-OE2 | 7±1Aa | 7±1AA | 16±2AA | 33±8AA | 3±1AA | 6±7Aa |
| ZmSTART1-OE3 | 6±1aa | 6±1Aa | 15±2Aa | 30±8AA | 2±1AA | 5±7aa |
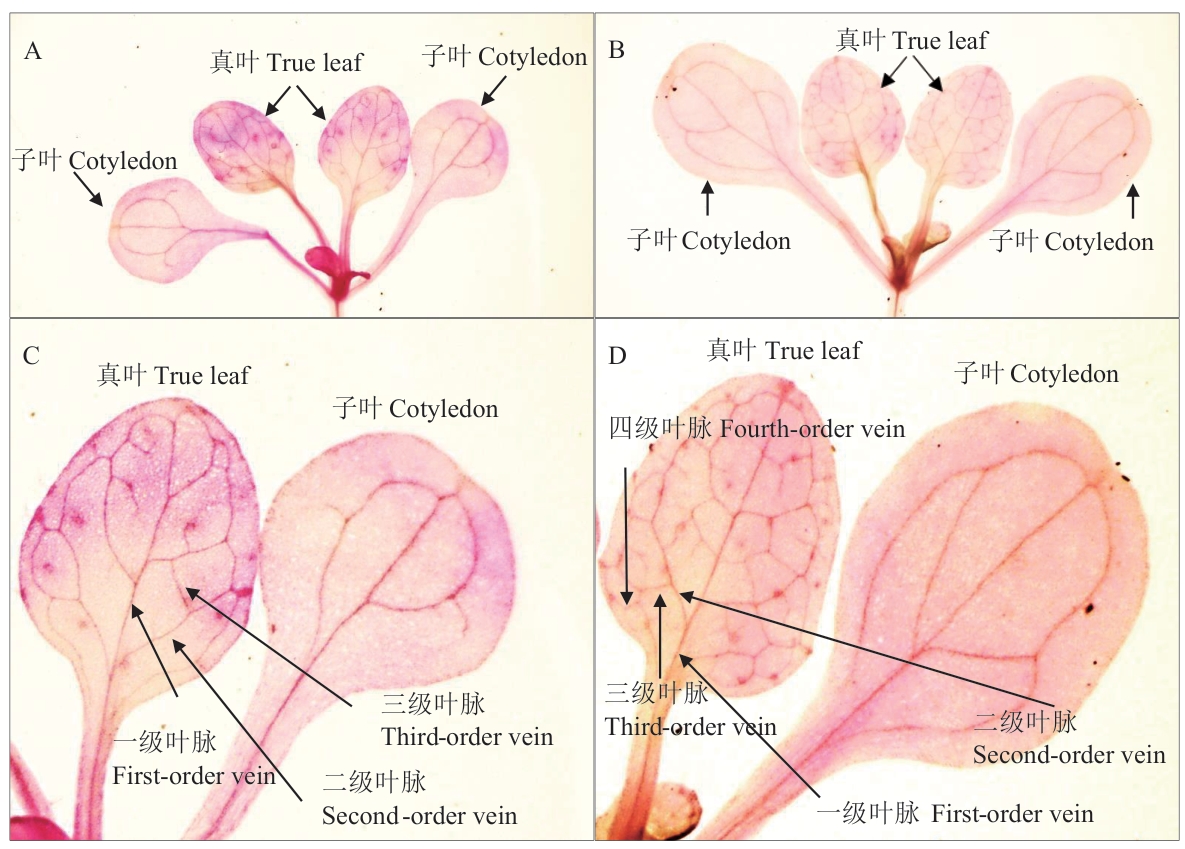
图7 过表达ZmSTART1转基因拟南芥叶片叶脉表型A和B:分别为野生型(A)和过表达ZmSTART1转基因拟南芥(B)子叶和真叶叶脉;C和D:分别为野生型(C)和过表达ZmSTART1转基因拟南芥(D)真叶各级叶脉
Fig. 7 Phenotype of the of ZmSTART1-overexpressed transgenic A.thaliana leaf veinA and B: The cotyledon and true leaf veins of wild type (A) and ZmSTART1-overexpressed transgenic Arabidopsis (B) respectively. C and D: The true leaf veins at all levels of wild type (C) and ZmSTART1-overexpressed transgenic Arabidopsis (D) respectively

图8 过表达ZmSTART1转基因拟南芥茎横切面显微结构A和B:分别为野生型(A)和过表达ZmSTART1转基因拟南芥(B)茎横切面显微结构;C和D:分别为野生型(C)和过表达ZmSTART1转基因拟南芥(D)茎维管束
Fig. 8 Stem cross seetional microstructure of transgenie A. thaliana overexpressing ZmSTART1A and B: Stem cross seetional microstructure of wild type (A) and ZmSTART1-overexpressed transgenic Arabidopsis (B) respectively. C and D: The vascular bundle of wild type (C) and ZmSTART1-overexpressed transgenic Arabidopsis (D) respectively
| 1 | Liao SY, Yan J, Xing HK, et al. Genetic basis of vascular bundle variations in rice revealed by genome-wide association study [J]. Plant Sci, 2021, 302: 110715. |
| 2 | Zheng YX, Hou P, Zhu LY, et al. Genome-wide association study of vascular bundle-related traits in maize stalk [J]. Front Plant Sci, 2021, 12: 699486. |
| 3 | Du Q, Wang HZ. The role of HD-ZIP III transcription factors and miR165/166 in vascular development and secondary cell wall formation [J]. Plant Signal Behav, 2015, 10(10): e1078955. |
| 4 | McConnell JR, Emery J, Eshed Y, et al. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots [J]. Nature, 2001, 411(6838): 709-713. |
| 5 | Zhong RQ, Ye ZH. Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels [J]. Plant Cell Physiol, 2004, 45(4): 369-385. |
| 6 | Fei C, Geng X, Xu ZJ, et al. Multiple areas investigation reveals the genes related to vascular bundles in rice [J]. Rice (N Y), 2019, 12(1): 17. |
| 7 | Fujita D, Trijatmiko KR, Tagle AG, et al. NAL1 allele from a rice Landrace greatly increases yield in modern indica cultivars [J]. Proc Natl Acad Sci USA, 2013, 110(51): 20431-20436. |
| 8 | Qi J, Qian Q, Bu QY, et al. Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport [J]. Plant Physiol, 2008, 147(4): 1947-1959. |
| 9 | Terao T, Nagata K, Morino K, et al. A gene controlling the number of primary Rachis branches also controls the vascular bundle formation and hence is responsible to increase the harvest index and grain yield in rice [J]. Theor Appl Genet, 2010, 120(5): 875-893. |
| 10 | Trieu AT, Burleigh SH, Kardailsky IV, et al. Transformation of Medicago truncatula via infiltration of seedlings or flowering plants with Agrobacterium [J]. Plant J, 2000, 22(6): 531-541. |
| 11 | Tsujishita Y, Hurley JH. Structure and lipid transport mechanism of a StAR-related domain [J]. Nat Struct Biol, 2000, 7(5): 408-414. |
| 12 | Clark BJ, Wells J, King SR, et al. The purification, cloning, and expression of a novel luteinizing hormone-induced mitochondrial protein in MA-10 mouse Leydig tumor cells. Characterization of the steroidogenic acute regulatory protein (StAR) [J]. J Biol Chem, 1994, 269(45): 28314-28322. |
| 13 | Waterman MR. A rising StAR: an essential role in cholesterol transport [J]. Science, 1995, 267(5205): 1780-1781. |
| 14 | Stocco DM. An update on the mechanism of action of the Steroidogenic Acute Regulatory (StAR) protein [J]. Exp Clin Endocrinol Diabetes, 1999, 107(4): 229-235. |
| 15 | de Brouwer APM, Westerman J, Kleinnijenhuis A, et al. Clofibrate-induced relocation of phosphatidylcholine transfer protein to mitochondria in endothelial cells [J]. Exp Cell Res, 2002, 274(1): 100-111. |
| 16 | Garajová K, Zimmermann M, Petrenčáková M, et al. The molten-globule residual structure is critical for reflavination of glucose oxidase [J]. Biophys Chem, 2017, 230: 74-83. |
| 17 | Soccio RE, Adams RM, Maxwell KN, et al. Differential gene regulation of StarD4 and StarD5 cholesterol transfer proteins. Activation of StarD4 by sterol regulatory element-binding protein-2 and StarD5 by endoplasmic reticulum stress [J]. J Biol Chem, 2005, 280(19): 19410-19418. |
| 18 | LaVoie HA, Whitfield NE, Shi B, et al. STARD6 is expressed in steroidogenic cells of the ovary and can enhance de novo steroidogenesis [J]. Exp Biol Med, 2014, 239(4): 430-435. |
| 19 | Al Haddad M, El-Rif R, Hanna S, et al. Differential regulation of rho GTPases during lung adenocarcinoma migration and invasion reveals a novel role of the tumor suppressor StarD13 in invadopodia regulation [J]. Cell Commun Signal, 2020, 18(1): 144. |
| 20 | Prigge MJ, Otsuga D, Alonso JM, et al. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development [J]. Plant Cell, 2005, 17(1): 61-76. |
| 21 | Rao RP, Yuan CQ, Allegood JC, et al. Ceramide transfer protein function is essential for normal oxidative stress response and lifespan [J]. Proc Natl Acad Sci U S A, 2007, 104(27): 11364-11369. |
| 22 | Alpy F, Tomasetto C. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals [J]. J Cell Sci, 2005, 118(Pt 13): 2791-2801. |
| 23 | Byrne ME. Shoot meristem function and leaf polarity: the role of class III HD-ZIP genes [J]. PLoS Genet, 2006, 2(6): e89. |
| 24 | Baima S, Possenti M, Matteucci A, et al. The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems [J]. Plant Physiol, 2001, 126(2): 643-655. |
| [1] | 王涛, 胡社伟, 张宇, 邓文文, 尚春缘, 王婉艺. 玉米籽粒淀粉生物合成及调控因素研究进展[J]. 生物技术通报, 2025, 41(3): 1-13. |
| [2] | 任鑫茹, 赵宏璐, 李雅静, 刘荣军, 曾凡力, 王钦宏, 王震. 混菌低温发酵对玉米秸秆黄贮饲料品质的影响[J]. 生物技术通报, 2025, 41(3): 330-342. |
| [3] | 田栩瑞, 霍信屹, 郭云涵, 向林, 产祝龙, 王艳平. 百合LoSAUR10基因的表达特征及功能分析[J]. 生物技术通报, 2025, 41(1): 221-229. |
| [4] | 林彤, 袁程, 董陈文华, 曾孟琼, 杨燕, 毛自朝, 林春. 藜麦配子发育相关基因CqSTK的筛选及功能分析[J]. 生物技术通报, 2024, 40(8): 83-94. |
| [5] | 任晓敏, 云岚, 艾芊, 赵乔. 新麦草异戊烯基转移酶PjIPT基因的功能验证[J]. 生物技术通报, 2024, 40(7): 207-215. |
| [6] | 王秋月, 段鹏亮, 李海笑, 刘宁, 曹志艳, 董金皋. 玉米大斑病菌cDNA文库的构建及转录因子StMR1互作蛋白的筛选[J]. 生物技术通报, 2024, 40(6): 281-289. |
| [7] | 胡锦锦, 李素贞, 马旭辉, 柳小庆, 谢珊珊, 江海洋, 陈茹梅. 玉米花青素生物合成代谢调控[J]. 生物技术通报, 2024, 40(6): 34-44. |
| [8] | 孙亚楠, 王春雪, 王欣, 杜秉海, 刘凯, 汪城墙. 萎缩芽孢杆菌CNY01的生防特性及其对玉米的抗盐促生作用[J]. 生物技术通报, 2024, 40(5): 248-260. |
| [9] | 李景艳, 周家婧, 袁媛, 苏晓艺, 乔文慧, 薛岩磊, 李国婧, 王瑞刚. 拟南芥AtiPGAM2基因参与非生物胁迫的响应[J]. 生物技术通报, 2024, 40(5): 215-224. |
| [10] | 陈春林, 李白雪, 李金玲, 杜清洁, 李猛, 肖怀娟. 甜瓜CmEPF基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(4): 130-138. |
| [11] | 王佳玮, 李晨, 刘建利, 周世杰, 易嘉敏, 杨谨源, 康鹏. 内生真菌接种方式对青贮玉米幼苗生长的影响[J]. 生物技术通报, 2024, 40(4): 189-202. |
| [12] | 张以恒, 刘家正, 王雪晨, 孙政哲, 薛雅郡, 汪沛, 韩华, 郑宏伟, 李晓娟. 基于超分辨成像增强对拟南芥内质网动态变化的研究[J]. 生物技术通报, 2024, 40(4): 67-76. |
| [13] | 胡伊娃, 陈露. 玉米野生种基因组研究进展及应用[J]. 生物技术通报, 2024, 40(3): 14-24. |
| [14] | 殷子薇, 红雨. 玫瑰红球菌NB1对玉米的耐盐促生效应及其全基因组研究[J]. 生物技术通报, 2024, 40(12): 193-207. |
| [15] | 王晶, 张晓磊, 白玉, 盛宇欣, 关海涛, 温洪涛. 不同玉米转化体通用PCR检测体系建立[J]. 生物技术通报, 2024, 40(12): 34-44. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||