








生物技术通报 ›› 2026, Vol. 42 ›› Issue (1): 125-138.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0560
龙林茜( ), 曾银萍, 王茜, 邓玉萍, 葛敏茜, 陈彦灼, 李鑫娟, 杨军(
), 曾银萍, 王茜, 邓玉萍, 葛敏茜, 陈彦灼, 李鑫娟, 杨军( ), 邹建(
), 邹建( )
)
收稿日期:2025-05-31
出版日期:2026-01-26
发布日期:2026-02-04
通讯作者:
邹建,男,博士,副教授,研究方向 :植物遗传与发育;E-mail: zoujian@cwnu.edu.cn作者简介:龙林茜,女,硕士研究生,研究方向 :植物遗传与发育;E-mail: longlinxi0212@163.com
基金资助:
LONG Lin-xi( ), ZENG Yin-ping, WANG Qian, DENG Yu-ping, GE Min-qian, CHEN Yan-zhuo, LI Xin-juan, YANG Jun(
), ZENG Yin-ping, WANG Qian, DENG Yu-ping, GE Min-qian, CHEN Yan-zhuo, LI Xin-juan, YANG Jun( ), ZOU Jian(
), ZOU Jian( )
)
Received:2025-05-31
Published:2026-01-26
Online:2026-02-04
摘要:
目的 探究向日葵(Helianthus annuus L.)HaGH3家族成员的理化特征和表达分析,以及其在向日葵花发育中的作用,为向日葵品种选育提供参考。 方法 基于向日葵基因组和转录组数据,利用生物信息学方法筛选鉴定HaGH3家族成员,并对蛋白理化性质、染色体定位、系统进化关系、保守基序、顺式作用元件和组织表达特征等进行分析。利用RT-qPCR分析HaGH3家族成员在野生型向日葵及其花分生组织转变与花器官发生缺陷突变体cb1中St2‒St8时期的表达差异,并对其在St5和St6时期不同花器官中的表达情况进行分析。 结果 向日葵HaGH3基因家族包含19个成员(HaGH3.1‒HaGH3.19),分散分布在12条染色体上,分属于2个亚家族,同一亚家族的成员具有相同的蛋白保守结构域。转录组数据分析显示,HaGH3.1、HaGH3.2、HaGH3.3、HaGH3.4、HaGH3.5、HaGH3.8、HaGH3.11、HaGH3.12、HaGH3.13、HaGH3.14和HaGH3.18等11个基因在向日葵花发育特定时期高水平表达。基于RT-qPCR的表达分析,显示HaGH3.3、HaGH3.4、HaGH3.8、HaGH3.13和HaGH3.14在WT的St2时期表达水平达到峰值,且其表达水平在WT和cb1中存在显著差异;HaGH3.1、HaGH3.2、HaGH3.5、HaGH3.12和HaGH3.18在WT花发育后期(St5‒St6时期)高水平表达,且具有较强的花器官特异性。顺式作用元件分析显示,11个花发育相关HaGH3基因启动子区域内含有大量生长素和赤霉素响应元件和MIKC_MADS识别元件。进一步分析结果显示,HaGH3.8、HaGH3.14和HaGH3.18受到ARF介导的生长素信号调节,HaGH3.1和HaGH3.8受到MYB和bZIP等赤霉素信号相关转录因子的调控,HaGH3.2、HaGH3.8、HaGH3.13、HaGH3.14和HaGH3.18受到花同源异型转录因子MIKC_MADS的调控。 结论 向日葵HaGH3.1和HaGH3.3等11个成员在向日葵花生长发育的调控中发挥重要作用,其中HaGH3.1、HaGH3.2、HaGH3.8、HaGH3.13、HaGH3.14和HaGH3.18在调控花发育过程中,受到生长素或赤霉素信号相关转录因子以及MIKC_MADS成员的调控。
龙林茜, 曾银萍, 王茜, 邓玉萍, 葛敏茜, 陈彦灼, 李鑫娟, 杨军, 邹建. 向日葵GH3基因家族鉴定及其在花发育中的功能分析[J]. 生物技术通报, 2026, 42(1): 125-138.
LONG Lin-xi, ZENG Yin-ping, WANG Qian, DENG Yu-ping, GE Min-qian, CHEN Yan-zhuo, LI Xin-juan, YANG Jun, ZOU Jian. Identification of Sunflower GH3 Gene Family and Analysis of Their Function in Flower Development[J]. Biotechnology Bulletin, 2026, 42(1): 125-138.
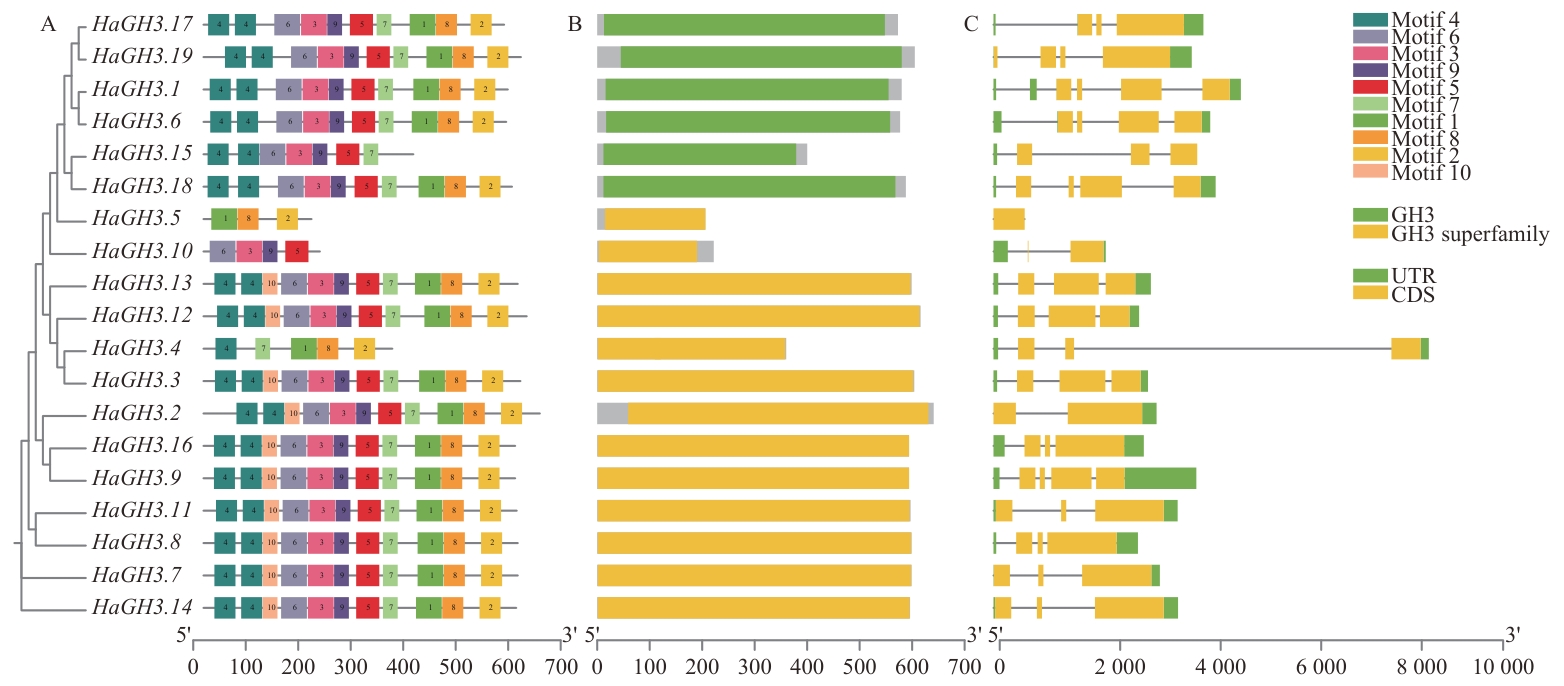
图2 HaGH3基因结构和蛋白保守基序分布A:HaGH3基因编码的蛋白保守基序;B:保守结构域图;C:基因结构图
Fig. 2 Gene structure and protein conserved motif distribution of HaGH3A: Conserved motif of protein encoded by HaGH3 gene. B: Conserved domain diagram. C: Gene structure map
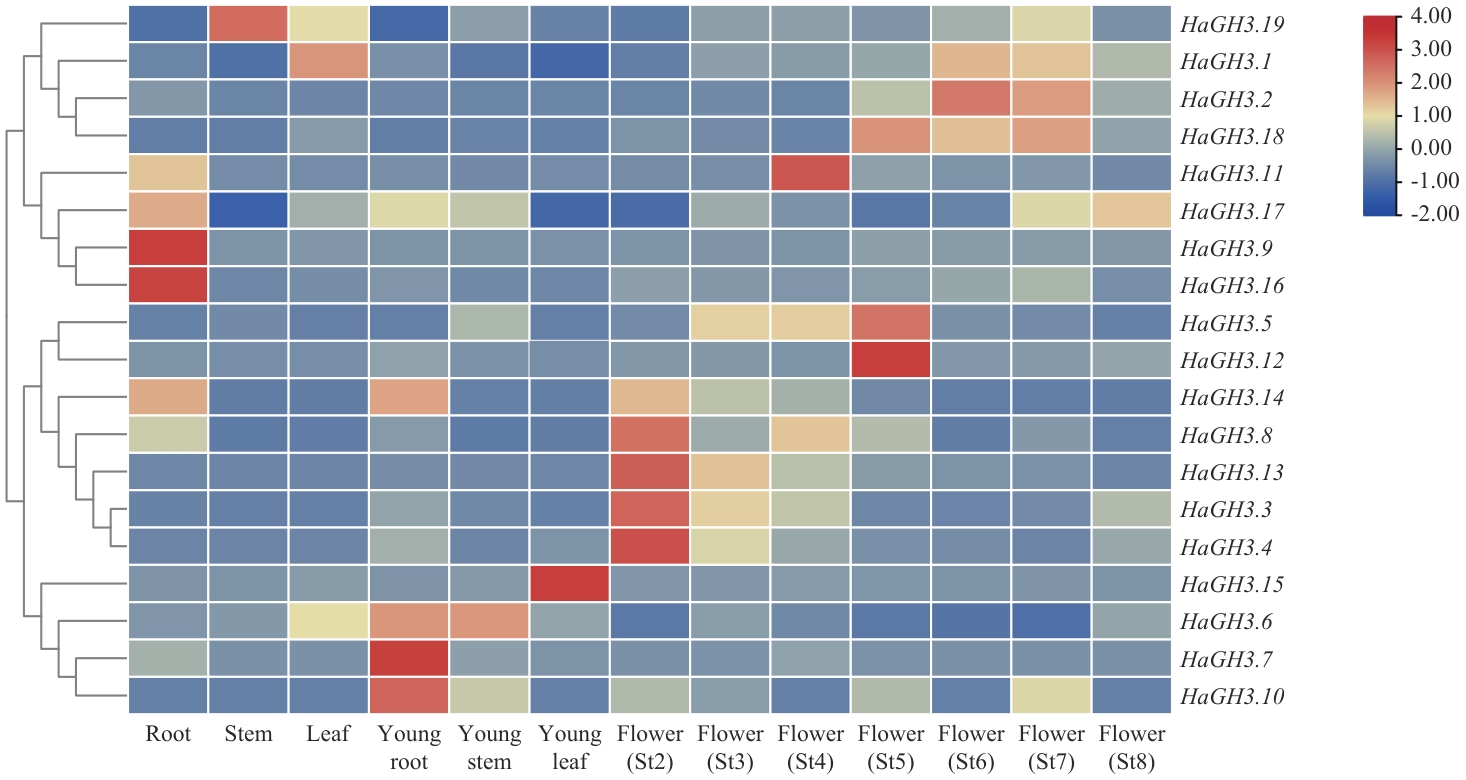
图3 HaGH3 家族基因在WT向日葵不同组织的表达分析St2:花原基起始期;St3:花器官开始形成期;St4:减数分裂期;St5:花粉成熟期;St6:小花开放期;St7:果实灌浆期;St8:种子饱满期;下同
Fig. 3 Expression analysis of HaGH3 family genes in different tissues of WT sunflowerSt2: Initiation phase of flower primordium. St3: Phase of flower organs starting formation. St4: Meiosis phase. St5: Phase of pollen maturity. St6: Floret opening phase. St7: Fruit filling phase. St8: Seed-full phase. The same below
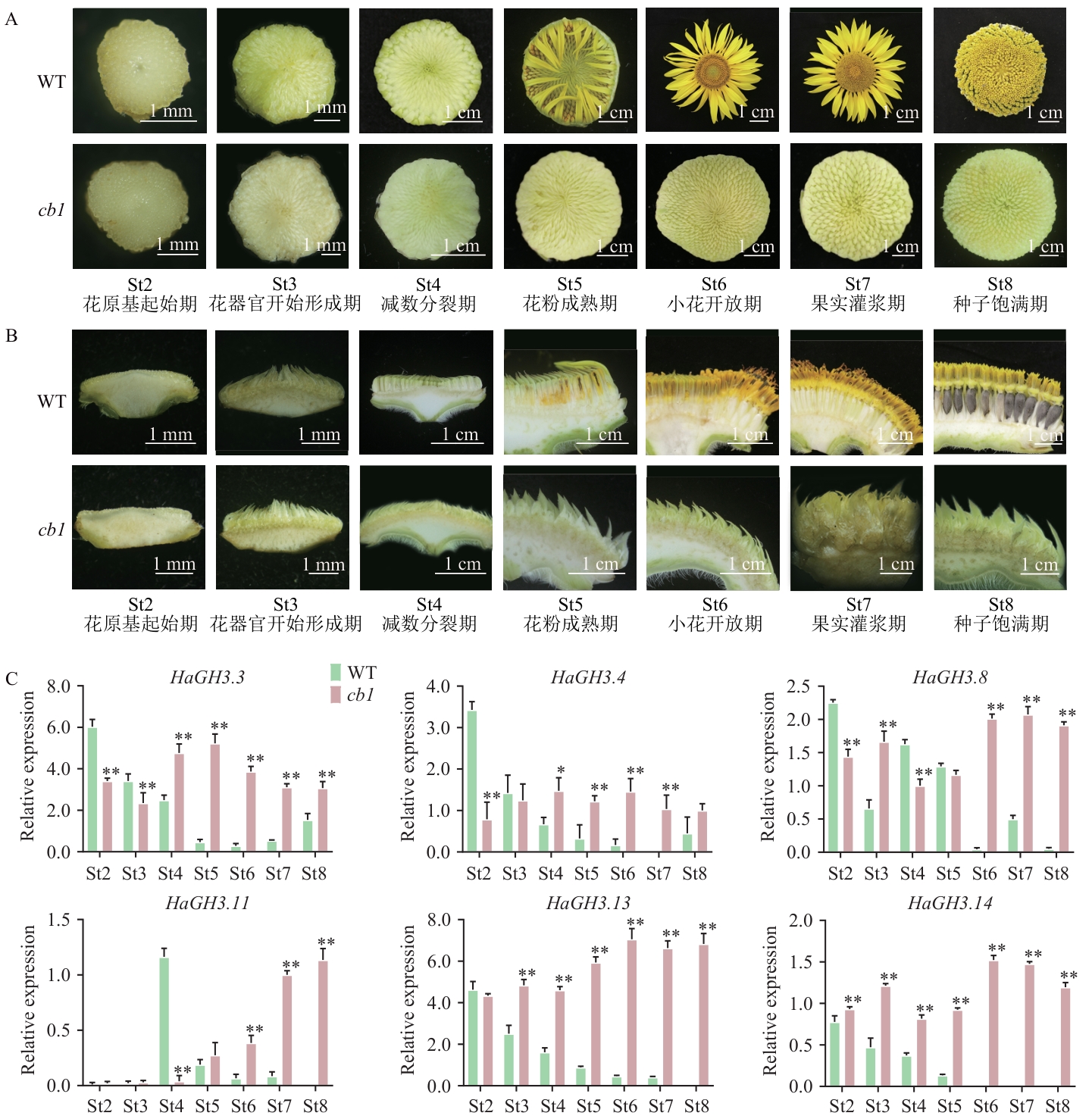
图4 HaGH3基因在野生型(WT)向日葵和cb1突变体中的差异表达分析A:WT向日葵和cb1突变体的花盘;B:WT向日葵和cb1突变体花序纵剖图;C:HaGH3基因在WT花和cb1突变体类花序组织(St2‒St8)的相对表达水平分析。数据为平均值±SD(n=3);*P<0.05,**P<0.01,下同
Fig. 4 Differential expression analysis of HaGH3 gene in WT sunflower and cb1 mutantA: Flower disc of WT sunflower and cb1 mutant. B: Longitudinal section of WT sunflower and cb1 mutant inflorescences. C: Relative expression of HaGH3 gene during inflorescence development of WT and cb1 mutants (St2‒St8). All data are mean ±SD (n=3). * P < 0.05, ** P < 0.01. The same below
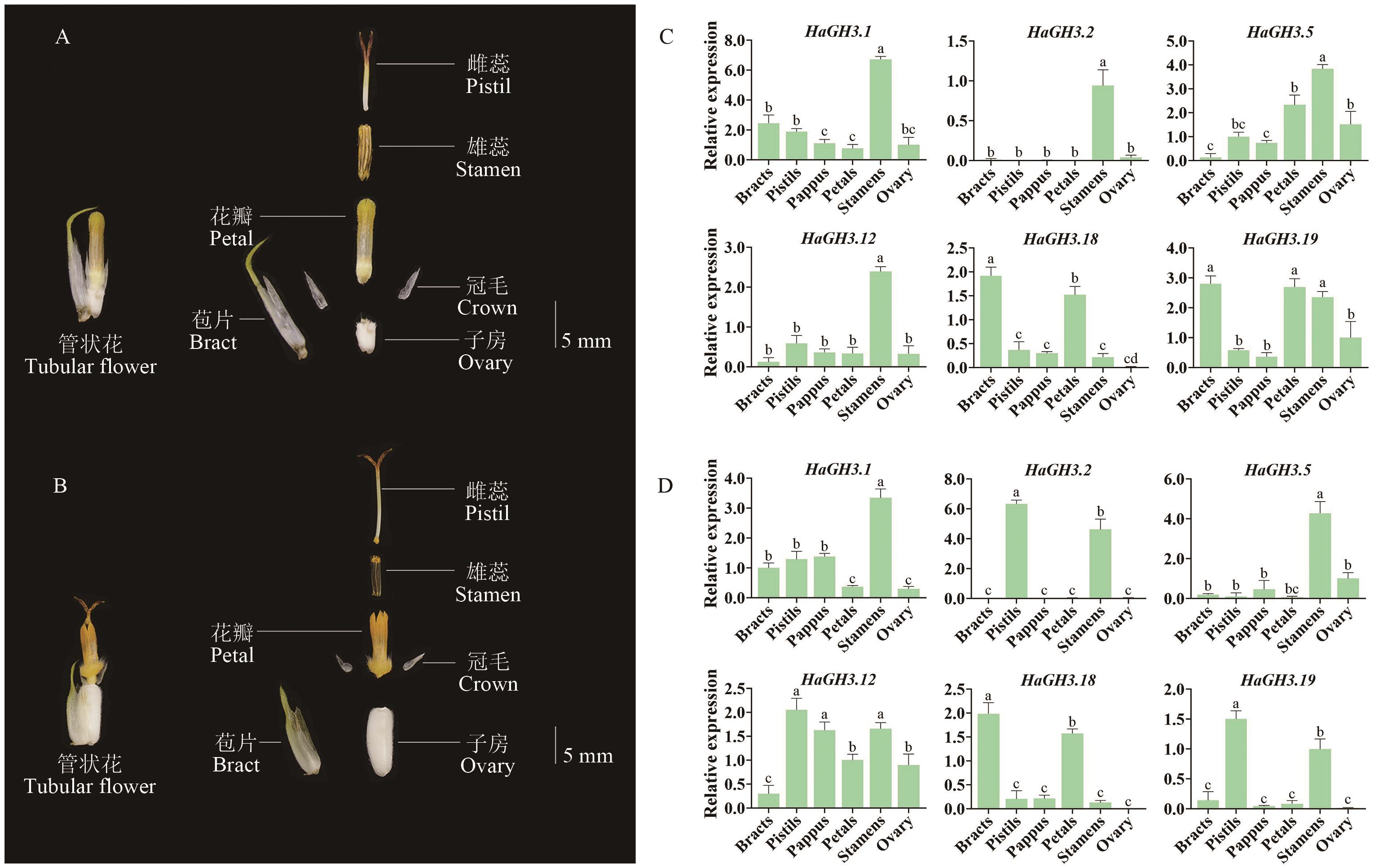
图5 HaGH3家族在不同时期花器官中的表达水平A:向日葵St5时期管状花解剖图;B:向日葵St6时期管状花解剖图;C:HaGH3基因家族在St5时期花器官中表达水平;D:在St6时期花器官中表达水平;数据以平均值±SD表示(n=3);不同小写字母表示组间差异具有统计学意义(P<0.05)
Fig. 5 Expressions of the HaGH3 family in flower organs at different phasesA: Anatomy of tubular flowers at St5 phase of sunflower. B: Anatomy of tubular flowers from the St6 phase of sunflower. C Expressions of sunflower GH3 gene family in flower organs at St5 phase. D: Expressions in flower organs at St6 phase. All data are mean ± SD (n=3). Groups marked with different lowercase letters indicate statistically significant differences (P < 0.05)
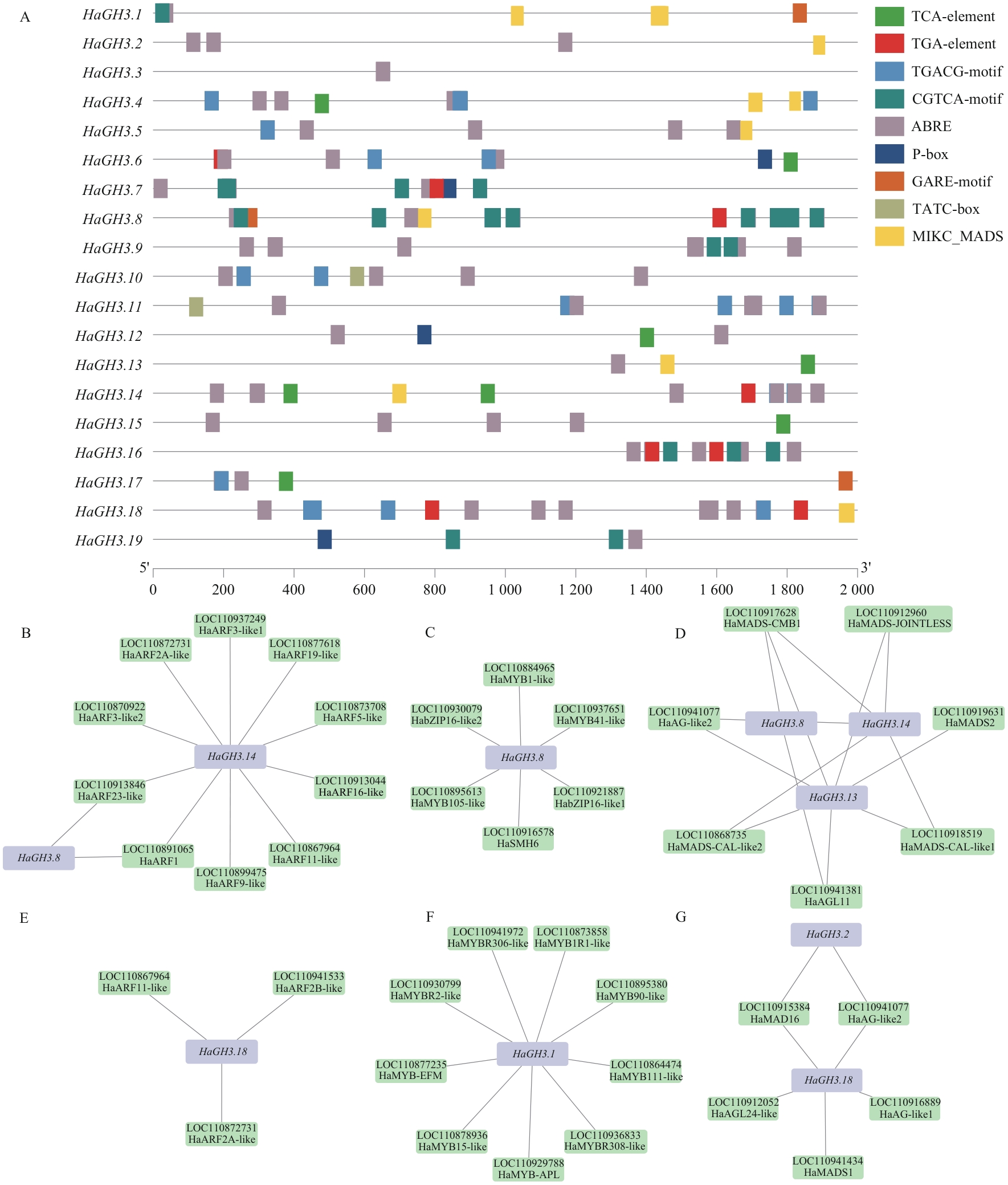
图6 HaGH3 基因启动子区顺式作用元件及其调控网络预测分析A:HaGH3基因顺式作用元件;B:HaGH3.8和HaGH3.14与ARF转录因子互作预测;C:HaGH3.8与MYB、bZIP转录因子互作预测;D:HaGH3.8、HaGH3.13和HaGH3.14基因与MIKC_MADS转录因子互作预测;E:HaGH3.18与ARF转录因子互作预测;F:HaGH3.1基因与MYB、bZIP转录因子互作预测;G:HaGH3.18基因与MIKC_MADS转录因子互作预测
Fig. 6 Prediction analysis of cis-acting elements and regulatory networks of the promoter region of HaGH3 geneA: Cis-acting element of the HaGH3 gene. B: Prediction of interaction between HaGH3.8 and HaGH3.14 with ARF transcription factors. C: Prediction of interaction between HaGH3.8 and MYB and bZIP transcription factors. D: Prediction of interaction between HaGH3.8, HaGH3.13 and HaGH3.14 genes with MIKC_MADS transcription factors. E: Prediction of interaction between HaGH3.18 and ARF transcription factors. F: Prediction of interaction between HaGH3.1 gene and MYB and bZIP transcription factors. G: Prediction of interaction between HaGH3.18 gene and MIKC_MADS transcription factors
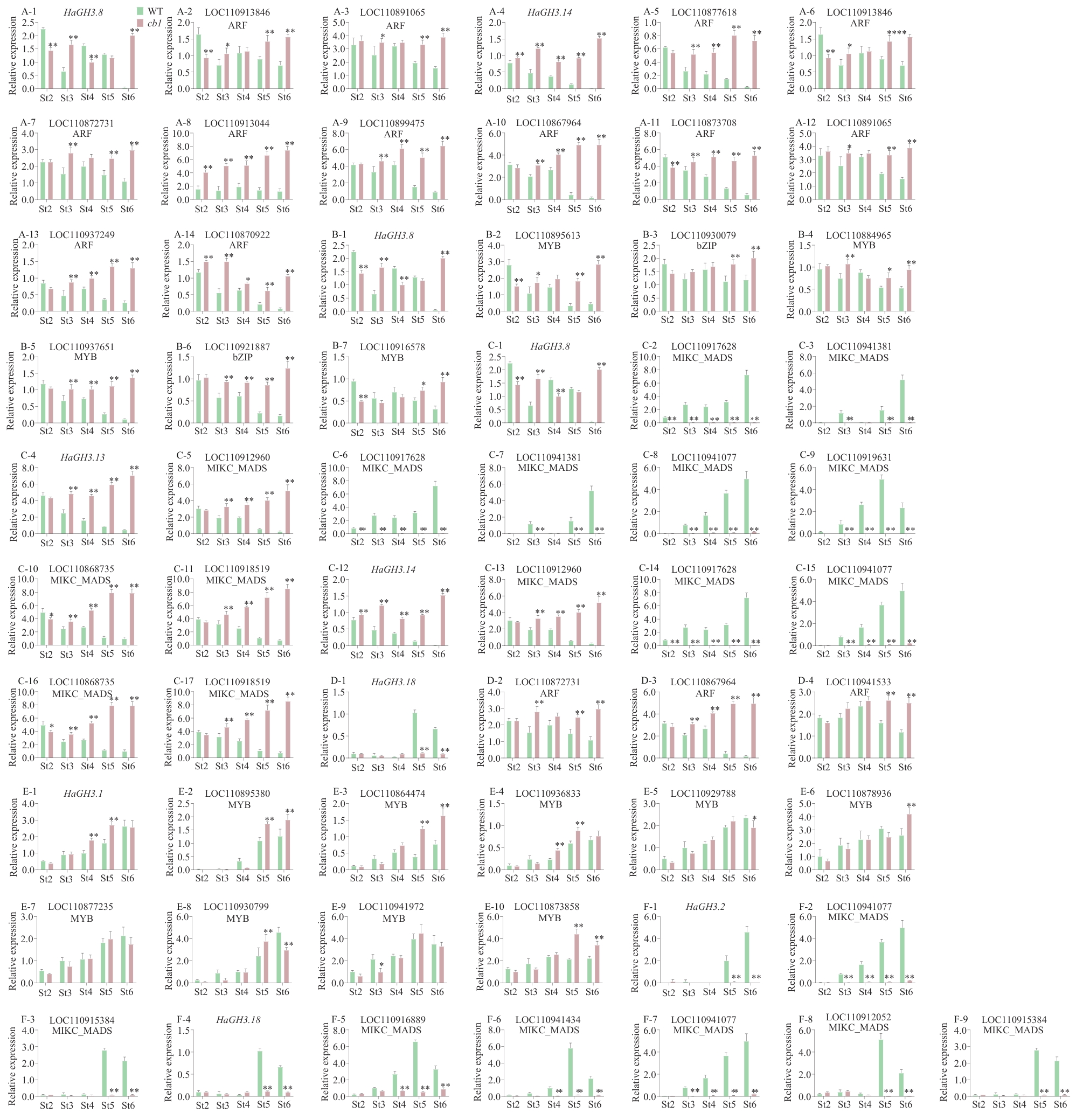
图7 参与花发育调控HaGH3基因及其候选调控转录因子的相对表达水平A:HaGH3.8和HaGH3.14与其潜在上游转录调控因子ARF的相对表达水平;B:HaGH3.8与其潜在上游转录调控因子MYB、bZIP的相对表达水平;C:HaGH3.8、HaGH3.13和HaGH3.14与其潜在上游转录调控因子MIKC_MADS的相对表达水平;D:HaGH3.18与其潜在上游转录调控因子ARF的相对表达水平;E:HaGH3.1与其潜在上游转录调控因子MYB、bZIP的相对表达水平;F:HaGH3.18与其潜在上游转录调控因子MIKC_MADS的相对表达水平
Fig. 7 Relative expressions of HaGH3 gene and its candidate transcription factors involved in the regulation of flower developmentA: Relative expressions of HaGH3.8 and HaGH3.14 and their potential upstream transcriptional regulators ARF. B: Relative expressions of HaGH3.8 and their potential upstream transcriptional regulators MYB and bZIP. C: Relative expressions of HaGH3.8, HaGH3.13 and HaGH3.14 and their potential upstream transcriptional regulators MIKC_MADS. D: Relative expressions of HaGH3.18 and its potential upstream transcriptional regulators ARF. E: Relative expressions of HaGH3.1 and its potential upstream transcriptional regulators MYB and bZIP. F: Relative expressions of HaGH3.18 and its potential upstream transcriptional regulatorsMIKC_MADS
| [1] | Wellmer F, Riechmann JL. Gene networks controlling the initiation of flower development [J]. Trends Genet, 2010, 26(12): 519-527. |
| [2] | Zhang C, Liu XF, Liu Y, et al. An integrated transcriptome and metabolome analysis reveals the gene network regulating flower development in Pogostemon cablin [J]. Front Plant Sci, 2023, 14: 1201486. |
| [3] | Zong J, Wang L, Zhu L, et al. A rice single cell transcriptomic atlas defines the developmental trajectories of rice floret and inflorescence meristems [J]. New Phytol, 2022, 234(2): 494-512. |
| [4] | Cheng XF, Li GF, Krom N, et al. Genetic regulation of flowering time and inflorescence architecture by MtFDa and MtFTa1 in Medicago truncatula [J]. Plant Physiol, 2021, 185(1): 161-178. |
| [5] | 金洲, 卢山, 江俊浩, 等. 园艺植物花芽分化影响因素及机理研究进展 [J]. 园艺学报, 2023, 50(5): 1151-1164. |
| Jin Z, Lu S, Jiang JH, et al. Research progress on influencing factors and mechanisms of flower bud differentiation in horticultural plants [J]. Acta Hortic Sin, 2023, 50(5): 1151-1164. | |
| [6] | 蒋存钰, 申彦华, 王义, 等. 生长素响应因子ARF研究进展 [J/OL]. 分子植物育种, 2022. . |
| Jiang CY, Shen YH, Wang Y, et al. Research progress of auxin response factor ARF [J/OL]. Mol Plant Breed, 2022. . | |
| [7] | Ma CH, Dang KT, Xie QK, et al. Over-expression of ZmIAA29, an AUX/IAA transcription factor, improved maize flowering time [J]. Agronomy, 2023, 13(8): 2028. |
| [8] | 园园, 恩和巴雅尔, 齐艳华. 植物GH3基因家族生物学功能研究进展 [J]. 植物学报, 2023, 58(5): 770-782. |
| Yuan Y, En H, Qi YH. Research advances in biological functions of GH3 gene family in plants [J]. Chin Bull Bot, 2023, 58(5): 770-782. | |
| [9] | Bao DF, Chang SQ, Li XD, et al. Advances in the study of auxin early response genes: Aux/IAA, GH3, and SAUR [J]. Crop J, 2024, 12(4): 964-978. |
| [10] | Staswick PE, Tiryaki I, Rowe ML. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation [J]. Plant Cell, 2002, 14(6): 1405-1415. |
| [11] | Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors [J]. Plant Mol Biol, 2002, 49(3/4): 373-385. |
| [12] | Jagadeeswaran G, Raina S, Acharya BR, et al. Arabidopsis GH3-LIKE DEFENSE GENE-1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae [J]. Plant J, 2007, 51(2): 234-246. |
| [13] | Nobuta RAO K. The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis [J]. Plant Physiol, 2007, 144(2): 1144-1156. |
| [14] | Woodward AW, Bartel B. Auxin: regulation, action, and interaction [J]. Ann Bot, 2005, 95(5): 707-735. |
| [15] | Nakazawa M, Yabe N, Ichikawa T, et al. DFL1 an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length [J]. Plant J, 2001, 25(2): 213-221. |
| [16] | Zhao SQ, Xiang JJ, Xue HW. Studies on the rice LEAF INCLINATION1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in leaf inclination control [J]. Mol Plant, 2013, 6(1): 174-187. |
| [17] | Du H, Wu N, Fu J, et al. A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice [J]. J Exp Bot, 2012, 63(18): 6467-6480. |
| [18] | Delfin JC, Kanno Y, Seo M, et al. AtGH3.10 is another jasmonic acid-amido synthetase in Arabidopsis thaliana [J]. Plant J, 2022, 110(4): 1082-1096. |
| [19] | Zhang SW, Li CH, Cao J, et al. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation [J]. Plant Physiol, 2009, 151(4): 1889-1901. |
| [20] | Singh VK, Jain M, Garg R. Genome-wide analysis and expression profiling suggest diverse roles of GH3 genes during development and abiotic stress responses in legumes [J]. Front Plant Sci, 2014, 5: 789. |
| [21] | 周苹, 唐冬英, 郭明, 等. 拟南芥GH3.9基因的过表达及其表型分析 [J]. 西北植物学报, 2015, 35(3): 454-458. |
| Zhou P, Tang DY, Guo M, et al. Overexpression and phenotype analysis of GH3.9 gene in Arabidopsis thaliana [J]. Acta Bot Boreali Occidentalia Sin, 2015, 35(3): 454-458. | |
| [22] | Yadav SR, Khanday I, Majhi BB, et al. Auxin-responsive OsMGH3, a common downstream target of OsMADS1 and OsMADS6, controls rice floret fertility [J]. Plant Cell Physiol, 2011, 52(12): 2123-2135. |
| [23] | Liu SY, Zhang CB, Guo F, et al. A systematical genome-wide analysis and screening of WRKY transcription factor family engaged in abiotic stress response in sweetpotato [J]. BMC Plant Biol, 2022, 22(1): 616. |
| [24] | Guo RP, Hu Y, Aoi Y, et al. Local conjugation of auxin by the GH3 amido synthetases is required for normal development of roots and flowers in Arabidopsis [J]. Biochem Biophys Res Commun, 2022, 589: 16-22. |
| [25] | Riemann M, Riemann M, Takano M. Rice JASMONATE RESISTANT 1 is involved in phytochrome and jasmonate signalling [J]. Plant Cell Environ, 2008, 31(6): 783-792. |
| [26] | Zhang SN, Wang SK, Xu YX, et al. The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1 [J]. Plant Cell Environ, 2015, 38(4): 638-654. |
| [27] | 周淑瑶, 李建明, 毛娟. AtGH3.17调控拟南芥生长素和油菜素甾醇的响应 [J]. 植物学报, 2023, 58(3): 373-384. |
| Zhou SY, Li JM, Mao J. AtGH3.17-mediated regulation of auxin and brassinosteroid response in Arabidopsis thaliana [J]. Chin Bull Bot, 2023, 58(3): 373-384. | |
| [28] | Mäkilä R, Wybouw B, Smetana O, et al. Gibberellins promote polar auxin transport to regulate stem cell fate decisions in cambium [J]. Nat Plants, 2023, 9(4): 631-644. |
| [29] | 邹礼平, 潘铖, 王梦馨, 等. 激素调控植物成花机理研究进展 [J]. 遗传, 2020, 42(8): 739-751. |
| Zou LP, Pan C, Wang MX, et al. Progress on the mechanism of hormones regulating plant flower formation [J]. Hereditas, 2020, 42(8): 739-751. | |
| [30] | Alejandra Freire-Rios, Keita Tanaka, Crespo, Isidro, et al. Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis [J]. Proc Natl Acad Sci U S A, 2020, 117(39): 24557-24566. |
| [31] | 宋松泉, 刘军, 黄荟, 等. 赤霉素代谢与信号转导及其调控种子萌发与休眠的分子机制 [J]. 中国科学: 生命科学, 2020, 50(6): 599-615. |
| Song SQ, Liu J, Huang H, et al. Gibberellin metabolism and signal transduction and its molecular mechanism of regulating seed germination and dormancy [J]. Sci China Ser C, 2020, 50(6): 599-615. | |
| [32] | Hernández-García J, Briones-Moreno A, Blázquez MA. Origin and evolution of gibberellin signaling and metabolism in plants [J]. Semin Cell Dev Biol, 2021, 109: 46-54. |
| [33] | Tsuji H, Aya K, Ueguchi-Tanaka M, et al. GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers [J]. Plant J, 2006, 47(3): 427-444. |
| [34] | Iven T, Strathmann A, Böttner S, et al. Homo- and heterodimers of tobacco bZIP proteins counteract as positive or negative regulators of transcription during pollen development [J]. Plant J, 2010, 63(1): 155-166. |
| [35] | 王莹, 穆艳霞, 王锦. MADS-box基因家族调控植物花器官发育研究进展 [J]. 浙江农业学报, 2021, 33(6): 1149-1158. |
| Wang Y, Mu YX, Wang J. Research progress of floral development regulation by MADS-box gene family [J]. Acta Agric Zhejiangensis, 2021, 33(6): 1149-1158. |
| [1] | 曾厅, 张兰, 罗睿. 转录因子MpR2R3-MYB17调控地钱胞芽发育的功能研究[J]. 生物技术通报, 2026, 42(1): 208-217. |
| [2] | 张驰昊, 刘晋囡, 晁跃辉. 蒺藜苜蓿bZIP转录因子MtbZIP29的克隆及功能分析[J]. 生物技术通报, 2026, 42(1): 241-250. |
| [3] | 吴翠翠, 陈登科, 兰刚, 夏芝, 李朋波. 花生转录因子AhHDZ70的生物信息学分析及耐盐耐旱性研究[J]. 生物技术通报, 2026, 42(1): 198-207. |
| [4] | 陈强, 于璎霏, 张颖, 张冲. 茉莉酸甲酯对薄皮甜瓜‘绿宝石’采后冷害的调控[J]. 生物技术通报, 2025, 41(9): 105-114. |
| [5] | 王斌, 林冲, 袁晓, 蒋园园, 王玉昆, 肖艳辉. bHLH转录因子UNE10克隆及其在丁香罗勒挥发性化合物合成调控中的功能[J]. 生物技术通报, 2025, 41(9): 207-218. |
| [6] | 黄诗宇, 田姗姗, 杨天为, 高曼熔, 张尚文. 赤苍藤WRI1基因家族的全基因组鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(8): 242-254. |
| [7] | 李开杰, 吴瑶, 李丹丹. 红花CtbHLH128基因克隆及调控干旱胁迫应答功能研究[J]. 生物技术通报, 2025, 41(8): 234-241. |
| [8] | 曾丹, 黄园, 王健, 张艳, 刘庆霞, 谷荣辉, 孙庆文, 陈宏宇. 铁皮石斛bZIP转录因子家族全基因组鉴定及表达分析[J]. 生物技术通报, 2025, 41(8): 197-210. |
| [9] | 高婧, 陈益存, 高暝, 赵耘霄, 汪阳东. 植物单宁合成调控及其对环境的响应机制[J]. 生物技术通报, 2025, 41(7): 49-59. |
| [10] | 牛景萍, 赵婧, 郭茜, 王书宏, 赵晋忠, 杜维俊, 殷丛丛, 岳爱琴. 基于WGCNA鉴定大豆抗大豆花叶病毒NAC转录因子及其诱导表达分析[J]. 生物技术通报, 2025, 41(7): 95-105. |
| [11] | 李霞, 张泽伟, 刘泽军, 王楠, 郭江波, 辛翠花, 张彤, 简磊. 马铃薯转录因子StMYB96的克隆及功能研究[J]. 生物技术通报, 2025, 41(7): 181-192. |
| [12] | 龚钰涵, 陈兰, 尚方慧子, 郝灵颖, 刘硕谦. 茶树TRB基因家族鉴定及表达模式分析[J]. 生物技术通报, 2025, 41(7): 214-225. |
| [13] | 魏雨佳, 李岩, 康语涵, 弓晓楠, 杜敏, 涂岚, 石鹏, 于子涵, 孙彦, 张昆. 白颖苔草CrMYB4基因的克隆和表达分析[J]. 生物技术通报, 2025, 41(7): 248-260. |
| [14] | 李小欢, 陈相宇, 陶麒宇, 朱玲, 唐铭, 姚银安, 汪丽君. PtoMYB61对毛白杨木质素合成及耐盐性的影响[J]. 生物技术通报, 2025, 41(6): 284-296. |
| [15] | 李锐, 胡婷, 陈树溦, 王尧, 王计平. 紫苏PfMYB80转录因子正向调控花青素的生物合成[J]. 生物技术通报, 2025, 41(6): 243-255. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||