








生物技术通报 ›› 2021, Vol. 37 ›› Issue (1): 2-14.doi: 10.13560/j.cnki.biotech.bull.1985.2020-0645
收稿日期:2020-05-26
出版日期:2021-01-26
发布日期:2021-01-15
作者简介:代文双,女,硕士研究生,研究方向:生物信息的整合与模拟;E-mail: 基金资助:
DAI Wen-shuang( ), LIU Hui-yun, DU Qing-guo, ZOU Cheng(
), LIU Hui-yun, DU Qing-guo, ZOU Cheng( ), WANG Ke(
), WANG Ke( )
)
Received:2020-05-26
Published:2021-01-26
Online:2021-01-15
摘要:
主要研究组蛋白去乙酰化酶抑制剂(HDACi)与染色质状态以及CRISPR/Cas9的编辑效率之间的关系。利用不同浓度的尼克酰胺(0,2.5和5 mmol/L)和丁酸钠(0,5和10 mmol/L)对小麦幼苗处理7 d和14 d,结果显示丁酸钠处理会抑制幼苗的生长,而尼克酰胺对幼苗影响较小。对尼克酰胺处理的小麦幼苗进行转录组测序,发现了一些有利于促进染色质状态开放的基因:6个甲基转移酶合成通路基因。此外对未发生编辑的TaAGO4a基因编辑转基因小麦材料的T2代进行尼克酰胺处理,结果显示5 mmol/L处理14 d时检测到1株3A和3B基因组均杂合编辑的植株,编辑效率从0提高到8.3%,其它处理组和对照组均没有检测到编辑。本研究证明尼克酰胺确实可以提高小麦基因编辑效率,为提高小麦基因编辑效率提供了一种新策略。
代文双, 刘会云, 杜庆国, 邹枨, 王轲. 组蛋白去乙酰化酶抑制剂(HDACi)对小麦基因编辑效率的影响及转录组学分析[J]. 生物技术通报, 2021, 37(1): 2-14.
DAI Wen-shuang, LIU Hui-yun, DU Qing-guo, ZOU Cheng, WANG Ke. Effect of Histone Deacetylase Inhibitor(HDACi)on CRISPR Editing Efficiency of Wheat and Transcriptomics Analysis[J]. Biotechnology Bulletin, 2021, 37(1): 2-14.
| 引物名称 | 引物序列(5'-3') |
|---|---|
| AGO4a486F | AGACTTTGCCTATGATGGTGA |
| AGO4a1404R | AAATCTTACCTTAGCCCAATCA |
| Ago4aA1629R | ACTTACATACAACAAATGGCGATATA |
| Ago4aB749F | GAAACGACAGTCCAGGAAATG |
| Ago4aD698F | GTTGCTCATGTAACTCTGTTGTTG |
表1 PCR引物序列表
| 引物名称 | 引物序列(5'-3') |
|---|---|
| AGO4a486F | AGACTTTGCCTATGATGGTGA |
| AGO4a1404R | AAATCTTACCTTAGCCCAATCA |
| Ago4aA1629R | ACTTACATACAACAAATGGCGATATA |
| Ago4aB749F | GAAACGACAGTCCAGGAAATG |
| Ago4aD698F | GTTGCTCATGTAACTCTGTTGTTG |
| 样本 | 非冗余 序列数 | Q20/% | Q30/% | GC/% | 比对率/% |
|---|---|---|---|---|---|
| ctrl7d_1 | 59882027 | 97.08 | 93.21 | 56.72 | 91.21 |
| ctrl7d_2 | 46792049 | 97.12 | 93.43 | 56.84 | 91.71 |
| t7d-2.5_1 | 51954951 | 96.74 | 92.69 | 57.80 | 90.85 |
| t7d-2.5_2 | 46624524 | 96.50 | 92.18 | 57.68 | 90.57 |
| t7d-5_1 | 43606982 | 96.87 | 92.91 | 57.30 | 91.48 |
| t7d-5_2 | 44386513 | 97.08 | 93.08 | 57.24 | 91.89 |
| ctrl14d _1 | 47377531 | 97.23 | 93.32 | 57.09 | 91.90 |
| ctrl14d _2 | 62520474 | 97.15 | 93.33 | 58.19 | 92.52 |
| t14d-2.5_1 | 44413018 | 96.83 | 92.58 | 57.21 | 91.76 |
| t14d-2.5_2 | 65941651 | 96.97 | 92.94 | 57.65 | 91.87 |
| t14d-5_1 | 48123569 | 96.92 | 93.16 | 58.24 | 90.87 |
| t14d-5_2 | 50461326 | 97.23 | 93.39 | 58.17 | 92.49 |
表2 测序数据质控与比对情况统计
| 样本 | 非冗余 序列数 | Q20/% | Q30/% | GC/% | 比对率/% |
|---|---|---|---|---|---|
| ctrl7d_1 | 59882027 | 97.08 | 93.21 | 56.72 | 91.21 |
| ctrl7d_2 | 46792049 | 97.12 | 93.43 | 56.84 | 91.71 |
| t7d-2.5_1 | 51954951 | 96.74 | 92.69 | 57.80 | 90.85 |
| t7d-2.5_2 | 46624524 | 96.50 | 92.18 | 57.68 | 90.57 |
| t7d-5_1 | 43606982 | 96.87 | 92.91 | 57.30 | 91.48 |
| t7d-5_2 | 44386513 | 97.08 | 93.08 | 57.24 | 91.89 |
| ctrl14d _1 | 47377531 | 97.23 | 93.32 | 57.09 | 91.90 |
| ctrl14d _2 | 62520474 | 97.15 | 93.33 | 58.19 | 92.52 |
| t14d-2.5_1 | 44413018 | 96.83 | 92.58 | 57.21 | 91.76 |
| t14d-2.5_2 | 65941651 | 96.97 | 92.94 | 57.65 | 91.87 |
| t14d-5_1 | 48123569 | 96.92 | 93.16 | 58.24 | 90.87 |
| t14d-5_2 | 50461326 | 97.23 | 93.39 | 58.17 | 92.49 |
| 处理 时间/d | GO terms | 基因 数目 | 类别 | 调控 方式 |
|---|---|---|---|---|
| 7 | nicotianamine synthase activity | 15 | MF | down |
| 7 | nicotianamine biosynthetic process | 15 | BP | down |
| 7 | O-methyltransferase activity | 6 | MF | down |
| 7 | transaminase activity | 6 | MF | up |
| 7 | cell division | 4 | BP | up |
表3 主要GO trems统计表
| 处理 时间/d | GO terms | 基因 数目 | 类别 | 调控 方式 |
|---|---|---|---|---|
| 7 | nicotianamine synthase activity | 15 | MF | down |
| 7 | nicotianamine biosynthetic process | 15 | BP | down |
| 7 | O-methyltransferase activity | 6 | MF | down |
| 7 | transaminase activity | 6 | MF | up |
| 7 | cell division | 4 | BP | up |
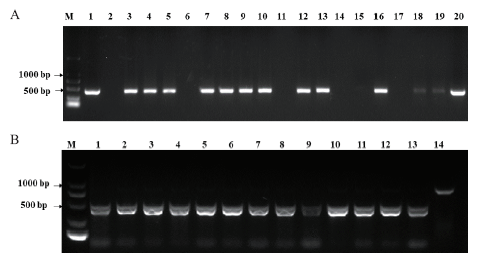
图6 T1代转基因植株bar基因及突变类型检测情况 A:T1代转基因植株bar基因PCR检测。M:2 kb DNA marker;1:阳性对照(质粒);2:阴性对照(水);3-8:QA147-1;9-14:QA167-2;15-19:QA167-7。B:利用通用引物对T1代转基因植株中TaAGO4a基因的PCR-RE检测,M:2 kb DNA marker;1-4:QA147-1;5-8:QA167-2;9-12:QA167-7;13:受体对照Fielder;14:未酶切的PCR产物
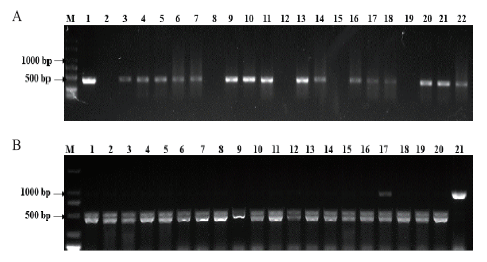
图7 14 d T2代转基因植株bar基因及突变类型检测情况 A:T2代转基因植株bar基因PCR检测。M:2 kb DNA marker;1:阳性对照(质粒);2:阴性对照(水);3-8:QA147-1;9-15:QA167-2;16-22:QA167-7。B:利用通用引物对T2代转基因植株TaAGO4a基因的PCR-RE检测。M:2 kb DNA marker;1-6:14-CK;7-13:14-2.5;14-19:14-5;20:受体对照Fielder;21:未酶切的PCR产物
| 样本 | 尼克酰胺浓度/(mmol·L-1) | 处理数 | 检测植株数 | Cas9阳性 | 编辑情况 | 编辑效率 |
|---|---|---|---|---|---|---|
| 7-CK | 0 | 14 | 19 | 12 | 0 | 0 |
| 7-2.5 | 2.5 | 14 | 32 | 18 | 0 | 0 |
| 7-5 | 5 | 14 | 18 | 12 | 0 | 0 |
| 14-CK | 0 | 14 | 19 | 12 | 0 | 0 |
| 14-2.5 | 2.5 | 14 | 32 | 18 | 0 | 0 |
| 14-5 | 5 | 14 | 18 | 12 | 1He | 8.3% |
表4 各处理编辑情况统计
| 样本 | 尼克酰胺浓度/(mmol·L-1) | 处理数 | 检测植株数 | Cas9阳性 | 编辑情况 | 编辑效率 |
|---|---|---|---|---|---|---|
| 7-CK | 0 | 14 | 19 | 12 | 0 | 0 |
| 7-2.5 | 2.5 | 14 | 32 | 18 | 0 | 0 |
| 7-5 | 5 | 14 | 18 | 12 | 0 | 0 |
| 14-CK | 0 | 14 | 19 | 12 | 0 | 0 |
| 14-2.5 | 2.5 | 14 | 32 | 18 | 0 | 0 |
| 14-5 | 5 | 14 | 18 | 12 | 1He | 8.3% |
| [1] | Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems[J]. Science, 2013,339(6121):819-823. |
| [2] |
Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology[J]. Nat Methods, 2013,10(10):957-963.
URL pmid: 24076990 |
| [3] |
Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes[J]. Cell, 2017,168(1-2):20-36.
URL pmid: 27866654 |
| [4] |
Bortesi L, Zhu C, Zischewski J, et al. Patterns of CRISPR/Cas9 activity in plants, animals and microbes[J]. Plant Biotechnology Journal, 2016,14(12):2203-2216.
doi: 10.1111/pbi.12634 URL pmid: 27614091 |
| [5] | Li W, Teng F, Li T, Zhou Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems[J]. Nature Biotechnology, 2013,31:684-686. |
| [6] |
马兴亮, 刘耀光. 植物CRISPR/Cas9基因组编辑系统与突变分析[J]. 遗传, 2016,38(2):118-125.
pmid: 26907775 |
|
Ma XL, Liu YG. CRISPR/Cas9-based genome editing systems and the analysis of targeted genome mutations in plants[J]. Hereditas, 2016,38(2):118-125.
doi: 10.16288/j.yczz.15-395 URL pmid: 26907775 |
|
| [7] |
Andersson M, Turesson H, Nicolia A, et al. Efficient targeted multiallelic mutagenesis in tetraploid potato(Solanum tuberosum)by transient CRISPR-Cas9 expression in protoplasts[J]. Plant Cell Reports, 2017,36(1):117-128.
doi: 10.1007/s00299-016-2062-3 URL pmid: 27699473 |
| [8] | Zhang Y, Liang Z, Zong Y, et al. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA[J]. Nature Communications, 2016,7:12617. |
| [9] | Angus WJ, Bonjean AP, Maarten VG. The world wheat book:a history of wheat breeding[M]. Paris:Lavoisier, 2001,2:122. |
| [10] | Bennet MD, Smith JB. Nuclear DNA amounts in angiosperms[J]. Philosophical Transactions of the Royal Society of London, 1991,334(1271):309-345. |
| [11] | 张正斌, 徐萍. 小麦基因组研究进展[J]. 遗传, 2002,24(3):389-394. |
| Zhang ZB, Xu P. Reviewed on wheat genome[J]. Hereditas, 2002,24(3):389-394. | |
| [12] |
Appels R, Eversole K, Feuillet C, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome[J]. Science, 2018, 361(6403):eaar7191.
doi: 10.1126/science.aar7191 URL |
| [13] | 于海霞, 田纪春. 普通小麦B基因组的研究进展[J]. 分子植物育种, 2008(4):724-732. |
| Yu HX, Tian JC. Review of genome B in T. aestivum L.[J]. Molecular Plant Breeding, 2008(4):724-732. | |
| [14] | Lu YZ, Wang L, Yue H, et al. Comparative analysis of stowaway-like miniature inverted repeat transposable elements in wheat group 7 chromosomes:abundance, composition, and evolution[J]. Journal of Systematics and Evolution, 2014,52(6):747-749. |
| [15] | Ke W, Liu H, Ye X, et al. Generation of marker-free transgenic hexaploid wheat via an agrobacterium-mediated co-transformation strategy in commercial Chinese wheat varieties[J]. Plant Biotechnology Journal, 2017,15(2):614-623. |
| [16] |
Kornberg RD. Chromatin structure:a repeating unit of histones and DNA[J]. Science, 1974,184(4139):868-871.
doi: 10.1126/science.184.4139.868 URL pmid: 4825889 |
| [17] |
Richmond TJ, Finch JT, Rushton B, et al. Structure of the nucleosome core particle at 7 A resolution[J]. Nature, 1984,311(5986):532-537.
doi: 10.1038/311532a0 URL pmid: 6482966 |
| [18] | Campos EI, Reinberg D. Histones:annotating chromatin[J]. Annual Review of Genetics, 2009,43(1):559-599. |
| [19] |
Hinz JM, Laughery MF, Wyrick JJ. Nucleosomes inhibit cas9 endonuclease activity in vitro[J]. Biochemistry, 2015,54(48):7063-7066.
doi: 10.1021/acs.biochem.5b01108 URL pmid: 26579937 |
| [20] |
Horlbeck MA, Witkowsky LB, Guglielmi B, et al. Nucleosomes impede Cas9 access to DNA in vivo and in vitro[J]. eLife, 2016,5:e12677.
doi: 10.7554/eLife.12677 URL pmid: 26987018 |
| [21] |
Yarrington RM, Verma S, Schwartz S, et al. Nucleosomes inhibit target cleavage by CRISPR-Cas9 in vivo[J]. PNAS, 2018,115(38):9351-9358.
doi: 10.1073/pnas.1810062115 URL pmid: 30201707 |
| [22] |
Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome[J]. Nature, 2012,489(7414):75-82.
URL pmid: 22955617 |
| [23] |
Wu X, Scott DA, Kriz AJ, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells[J]. Nature Biotechnology, 2014,32(7):670-676.
doi: 10.1038/nbt.2889 URL pmid: 24752079 |
| [24] | Kuscu C, Arslan S, Singh R, et al. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease[J]. Nat Biotech, 2014,32(7):677-683. |
| [25] |
Chari R, Mali P, Moosburner M, et al. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach[J]. Nature Methods, 2015,12(9):823-826.
doi: 10.1038/nmeth.3473 URL pmid: 26167643 |
| [26] |
Liu G, Yin K, Zhang Q, et al. Modulating chromatin accessibility by transactivation and targeting proximal dsgRNAs enhances Cas9 editing efficiency in vivo[J]. Genome Biol, 2019,20(1):145.
doi: 10.1186/s13059-019-1762-8 URL pmid: 31349852 |
| [27] | Isaac RS, Jiang F, Doudna JA, et al. Nucleosome breathing and remodeling constrain CRISPR-Cas9 function[J]. eLife, 2016: 5e13450. |
| [28] |
Bustin M. Chromatin unfolding and activation by HMGN* chromosomal proteins[J]. Trends in Biochemical Sciences, 2001,26(7):431-437.
doi: 10.1016/s0968-0004(01)01855-2 URL pmid: 11440855 |
| [29] |
Kugler JE, Deng T, Bustin M. The HMGN family of chromatin-binding proteins:dynamic modulators of epigenetic processes[J]. Biochimica et Biophysica Acta-gene Regulatory Mechanisms, 2012,1819(7):652-656.
doi: 10.1016/j.bbagrm.2012.01.013 URL |
| [30] |
Gonzalez-romero R, Eiri'n-lo'pez JM, Ausio' J. Evolution of high mobility group nucleosome-binding proteins and its implications for vertebrate chromatin specialization[J]. Molecular Biology and Evolution, 2015,32(1):121-131.
URL pmid: 25281808 |
| [31] |
Klass J, Murphy FV, Fouts S, et al. The role of intercalating residues in chromosomal high-mobility-group protein DNA binding, bending and specificity[J]. Nucleic Acids Research, 2003,31(11):2852-2864.
doi: 10.1093/nar/gkg389 URL pmid: 12771212 |
| [32] |
Thomas JO, Stott K. H1 and HMGB1:Modulators of chromatin structure[J]. Biochem Soc Trans, 2012,40(2):341-346.
doi: 10.1042/BST20120014 URL pmid: 22435809 |
| [33] |
Harshman SW, Young NL, Parthun MR, et al. H1 histones:current perspectives and challenges[J]. Nucleic Acids Research, 2013,41(21):9593-9609.
URL pmid: 23945933 |
| [34] | Ding X, Seebeck T, Feng Y, et al. Improving CRISPR-Cas9 genome editing efficiency by fusion with chromatin-modulating peptides[J]. CRISPR Journal, 2019,2(1):51-63. |
| [35] |
Wolffe AP, Guschin D. Review:chromatin structural features and targets that regulate transcription[J]. Journal of Structural Biology, 2000,129(2-3):102-122.
doi: 10.1006/jsbi.2000.4217 URL pmid: 10806063 |
| [36] |
Cheung WL, Briggs SD, Allis CD. Acetylation and chromosomal functions[J]. Curr Opin Cell Biol, 2000,12(3):326-333.
doi: 10.1016/s0955-0674(00)00096-x URL pmid: 10801466 |
| [37] | Allfrey VG, Mirsky RF. Acetylation and methylation of histones and their possible role in the regulation of RNA synjournal[J]. PNAS, 1964,51(5):786-794. |
| [38] |
René D, Cassandra MB, Karmella AH. Manipulation of chromatin to enhance CRISPR activity[J]. Biorxiv, 2018. http://doiorg/10.1101/228601.
URL pmid: 33501432 |
| [39] |
Bertos NR, Wang AH, Yang XJ. Class II histone deacetylases:structure, function, and regulation[J]. Biochemistry & Cell Biology, 2001,79(3):243-252.
URL pmid: 11467738 |
| [40] | Li W, Mills AA. Architects of the genome:CHD dysfunction in cancer, developmental disorders and neurological syndromes[J]. Epigenomics, 2014,6(4):381-395. |
| [41] | Demetrious K, Kapazoglou A, Tondelli A, et al. Epigenetic chromatin modifiers in barley:I. cloning, mapping and expression analysis of the plant specific HD2 family of histone deacetylases from barley, during seed development and after hormonal treatment[J]. Physiologia Plantarum, 2009,136(3):358-368. |
| [42] | Lagacé M, Chantha SC, Major G, et al. Fertilization induces strong accumulation of a histone deacetylase(HD2)and of other chromatin-remodeling proteins in restricted areas of the ovules[J]. Plant Molecular Biology, 2003,53(6):759-769. |
| [43] | Busconi M, Reggi S, Fogher C, et al. Evidence of a sirtuin gene family in grapevine(Vitis vinifera L.)[J]. Plant Physiol Biochem, 2009,47(7):650-652. |
| [44] | Bourque S, Dutartre A, Hammoudi V, et al. Type-2 histone deacetylases as new regulators of elicitor-induced cell death in plants[J]. New Phytologist, 2011,192(1):127-139. |
| [45] | Davie JR. Inhibition of histone deacetylase activity by butyrate[J]. J Nutr, 2003,133(7):2485-2493. |
| [46] | Bitterman KJ, Anderson RM, Cohen HY, et al. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human Sirt1[J]. Journal of Biological Chemistry, 2002,277(47):45099-45107. |
| [47] | Landry J, Slama JT, Sternglanz R. Role of NAD(+)in the deacetylase activity of the Sir2-like proteins[J]. Biochem Biophys Res Commun, 2000,278(3):685-690. |
| [48] | Sung S, Amasino RM. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3[J]. Nature(London), 2004,427(6970):159-164. |
| [49] |
Bond ES, Donna M, Dennis BJ. Histone acetylation, vernalization insensitive 3, flowering locus c, and the vernalization response[J]. Molecular Plant, 2009,2(4):724-737.
URL pmid: 19825652 |
| [50] |
Deng W, Liu C, Pei Y, et al. Involvement of the histone acetyltransferase AtHAC1 in the regulation of flowering time via repression of flowering locus c in Arabidopsis[J]. Plant Physiology, 2007,143(4):1660-1668.
doi: 10.1104/pp.106.095521 URL pmid: 17416640 |
| [51] |
Han SK, Song JD, Noh YS, et al. Role of plan CBP/p300-like genes in the regulation of flowering time[J]. The Plant Journal, 2007,49(1):103-114.
doi: 10.1111/j.1365-313X.2006.02939.x URL pmid: 17144897 |
| [52] | Tiricz H, Nagy B, Ferenc G, et al. Relaxed chromatin induced by histone deacetylase inhibitors improves the oligonucleotide-directed gene editing in plant cells[J]. Journal of Plant Research, 2018,131(1):179-189. |
| [53] |
Bolger AM, Lohse M, Usadel B. Trimmomatic:a flexible trimmer for Illumina sequence data[J]. Bioinformatics, 2014,30(15):2114-2120.
URL pmid: 24695404 |
| [54] | Kim D, Langmead B, Salzberg SL. Hisat:a fast spliced aligner with low memory requirements[J]. Nature Methods, 2015,12(4):357-360. |
| [55] |
Pertea M, Kim D, Pertea GM, et al. Transcript-level expression analysis of RNA-seq experiments with Hisat, StringTie and Ballgown[J]. Nature Protocols, 2016,11(9):1650-1667.
doi: 10.1038/nprot.2016.095 URL pmid: 27560171 |
| [56] | Pertea M, Pertea GM, Antonescu CM, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads[J]. Nature Biotechnology, 2015,33(3):290-295. |
| [57] |
Anders S, Paul TP, Huber W, et al. HTSeq--a Python framework to work with high-throughput sequencing data[J]. Bioinformatics, 2015,31(2):166-169.
URL pmid: 25260700 |
| [58] |
Li W, Mills AA. Architects of the genome:CHD dysfunction in cancer, developmental disorders and neurological syndromes[J]. Epigenomics, 2014,6(4):381-395.
doi: 10.2217/epi.14.31 URL |
| [59] |
Jones PL, Veenstra GJ, Wade PA, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription[J]. Nature Genetics, 1998,19(2):187-191.
doi: 10.1038/561 URL pmid: 9620779 |
| [60] |
Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex[J]. Nature, 1998,393(6683):386-389.
doi: 10.1038/30764 URL pmid: 9620804 |
| [61] | Turek-plewa J, Jagodziński PP. The role of mammalian DNA methyltransferases in the regulation of gene expression[J]. Cellular & Molecular Biology Letters, 2005,10(4):631-647. |
| [62] | Zhao Q, Rank G, Tan YT, et al. Prmt5-mediated methylation of histone H4r3 recruits Dnmt3a, coupling histone and DNA methylation in gene silencing[J]. Nature Structural & Molecular Biology, 2009,16(3):304-311. |
| [63] | Bachman KE, Rountree MR, Baylin SB. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin[J]. Journal of Biological Chemistry, 2001,276(34):32282-32287. |
| [64] | Kathleen RS, Nazaret RG, Anja G. Transcription restart establishes chromatin accessibility after DNA replication[J]. Molecular Cell, 2019,75(2):408-414. |
| [65] | Liu WZ, Xie XR, Ma XL, et al. DSDecode:A Web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations[J]. Molecular Plant, 2015,8(9):1431-1433. |
| [1] | 林红妍, 郭晓蕊, 刘迪, 李慧, 陆海. 转录组分析转录因子AtbHLH68调控细胞壁发育的分子机制[J]. 生物技术通报, 2023, 39(9): 105-116. |
| [2] | 温晓蕾, 李建嫄, 李娜, 张娜, 杨文香. 小麦叶锈菌与小麦互作的酵母双杂交cDNA文库构建与应用[J]. 生物技术通报, 2023, 39(9): 136-146. |
| [3] | 苗永美, 苗翠苹, 于庆才. 枯草芽孢杆菌BBs-27发酵液性质及脂肽对黄色镰刀菌的抑菌作用[J]. 生物技术通报, 2023, 39(9): 255-267. |
| [4] | 陈小玲, 廖东庆, 黄尚飞, 陈英, 芦志龙, 陈东. 利用CRISPR/Cas9系统改造酿酒酵母的研究进展[J]. 生物技术通报, 2023, 39(8): 148-158. |
| [5] | 杨玉梅, 张坤晓. 应用CRISPR/Cas9技术建立ERK激酶相分离荧光探针定点整合的稳定细胞株[J]. 生物技术通报, 2023, 39(8): 159-164. |
| [6] | 韩志阳, 贾子苗, 梁秋菊, 王轲, 唐华丽, 叶兴国, 张双喜. 二套小麦-簇毛麦染色体附加系苗期耐盐性及籽粒硒和叶酸的含量[J]. 生物技术通报, 2023, 39(8): 185-193. |
| [7] | 付钰, 贾瑞瑞, 何荷, 王良桂, 杨秀莲. 两种砧木楸树嫁接苗生长差异及转录组比较分析[J]. 生物技术通报, 2023, 39(8): 251-261. |
| [8] | 施炜涛, 姚春鹏, 魏文康, 王蕾, 房元杰, 仝钰洁, 马晓姣, 蒋文, 张晓爱, 邵伟. 利用CRISPR/Cas9技术构建MDH2敲除细胞株及抗呕吐毒素效应研究[J]. 生物技术通报, 2023, 39(7): 307-315. |
| [9] | 赵金玲, 安磊, 任晓亮. 单细胞转录组测序技术及其在秀丽隐杆线虫中的应用[J]. 生物技术通报, 2023, 39(6): 158-170. |
| [10] | 孔德真, 段震宇, 王刚, 张鑫, 席琳乔. 盐、碱胁迫下高丹草苗期生理特征及转录组学分析[J]. 生物技术通报, 2023, 39(6): 199-207. |
| [11] | 刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191. |
| [12] | 刘晓燕, 祝振亮, 史广宇, 华梓宇, 杨晨, 张涌, 刘军. 乳腺生物反应器的表达优化策略[J]. 生物技术通报, 2023, 39(5): 77-91. |
| [13] | 谢洋, 邢雨蒙, 周国彦, 刘美妍, 银珊珊, 闫立英. 黄瓜二倍体及其同源四倍体果实转录组分析[J]. 生物技术通报, 2023, 39(3): 152-162. |
| [14] | 扈丽丽, 林柏荣, 王宏洪, 陈建松, 廖金铃, 卓侃. 最短尾短体线虫转录组及潜在效应蛋白分析[J]. 生物技术通报, 2023, 39(3): 254-266. |
| [15] | 程静雯, 曹磊, 张艳敏, 叶倩, 陈敏, 谭文松, 赵亮. CHO细胞多基因工程改造策略的建立及应用[J]. 生物技术通报, 2023, 39(2): 283-291. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||