








生物技术通报 ›› 2023, Vol. 39 ›› Issue (2): 283-291.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0863
程静雯1( ), 曹磊1, 张艳敏1, 叶倩1, 陈敏2, 谭文松1, 赵亮1,2(
), 曹磊1, 张艳敏1, 叶倩1, 陈敏2, 谭文松1, 赵亮1,2( )
)
收稿日期:2022-07-14
出版日期:2023-02-26
发布日期:2023-03-07
作者简介:程静雯,女,硕士,研究方向:动物细胞与组织工程;E-mail:
CHENG Jing-wen1( ), CAO Lei1, ZHANG Yan-min1, YE Qian1, CHEN Min2, TAN Wen-song1, ZHAO Liang1,2(
), CAO Lei1, ZHANG Yan-min1, YE Qian1, CHEN Min2, TAN Wen-song1, ZHAO Liang1,2( )
)
Received:2022-07-14
Published:2023-02-26
Online:2023-03-07
摘要:
近年来,中国仓鼠卵巢(Chinese hamster ovary, CHO)细胞工程改造主要通过敲入或敲除基因来改变细胞某个单一功能,而敲入和敲除的基因往往无法在单次实验操作中同时发挥相应的功能,限制了多基因同步改造的应用。该研究选取细胞凋亡通路中抗凋亡蛋白即B淋巴细胞瘤-2(B-cell lymphoma-2, Bcl-2)基因为敲入基因、蛋白岩藻糖基化通路中岩藻糖合成关键酶即岩藻糖转移酶8(fucosyltransferase 8, FUT8)基因为敲除基因作为模型,利用CRISPR/Cas9技术建立定点同步敲入敲除基因编辑策略。利用该策略获得的单克隆细胞株Bcl-2蛋白过表达且FUT8蛋白酶功能缺失。经传代培养发现,由建立的定点同步敲入敲除基因编辑策略获得的细胞株在60 d内所编辑的基因表达稳定。相较于原野生型细胞,该细胞株表现出对血清剥夺的耐受度更高以及对死亡的抵抗能力更强。由此说明基于定点同步敲入敲除基因编辑策略具备一定可行性,可用于重组蛋白生产的CHO工程细胞株的构建。
程静雯, 曹磊, 张艳敏, 叶倩, 陈敏, 谭文松, 赵亮. CHO细胞多基因工程改造策略的建立及应用[J]. 生物技术通报, 2023, 39(2): 283-291.
CHENG Jing-wen, CAO Lei, ZHANG Yan-min, YE Qian, CHEN Min, TAN Wen-song, ZHAO Liang. Establishment and Application of Multigene Engineering Transformation Strategy for CHO Cells[J]. Biotechnology Bulletin, 2023, 39(2): 283-291.
| 寡核苷酸链名称Oligos name | DNA序列DNA sequences(5'-3') |
|---|---|
| FUT8-sg1F | CACCGATGGACTGGTTCCTGGCGT |
| FUT8-sg1 R | AAACACGCCAGGAACCAGTCCATC |
| FUT8-sg2 F | CACCGTAAAACAATAAGGTCCCCC |
| FUT8-sg2 R | AAACGGGGGACCTTATTGTTTTAC |
| FUT8-sg3 F | CACCGAGAAGGCCCTATTGATCAG |
| FUT8-sg3 R | AAACCTGATCAATAGGGCCTTCTC |
表1 sgRNA寡核苷酸链
Table 1 sgRNA oligos
| 寡核苷酸链名称Oligos name | DNA序列DNA sequences(5'-3') |
|---|---|
| FUT8-sg1F | CACCGATGGACTGGTTCCTGGCGT |
| FUT8-sg1 R | AAACACGCCAGGAACCAGTCCATC |
| FUT8-sg2 F | CACCGTAAAACAATAAGGTCCCCC |
| FUT8-sg2 R | AAACGGGGGACCTTATTGTTTTAC |
| FUT8-sg3 F | CACCGAGAAGGCCCTATTGATCAG |
| FUT8-sg3 R | AAACCTGATCAATAGGGCCTTCTC |
| 引物名称Primer name | DNA序列DNA sequence(5'-3') |
|---|---|
| FUT8-Exon1-F | AGAGTCATCACAGTATACCAGAGAG |
| FUT8-Exon1-R | GCCACTGCTTCTATATACTGATTCA |
| FUT8-Exon2-F | CATTCTCAGCTAGCCCTTATGATTA |
| FUT8-Exon2-R | TATGGAAGCCCAAATGAAGCACA |
表2 T7E I酶切实验引物
Table 2 PCR primers for T7E I enzymatic digestion assay
| 引物名称Primer name | DNA序列DNA sequence(5'-3') |
|---|---|
| FUT8-Exon1-F | AGAGTCATCACAGTATACCAGAGAG |
| FUT8-Exon1-R | GCCACTGCTTCTATATACTGATTCA |
| FUT8-Exon2-F | CATTCTCAGCTAGCCCTTATGATTA |
| FUT8-Exon2-R | TATGGAAGCCCAAATGAAGCACA |
| 目的 Purpose | 引物名称 Primer name | DNA序列 DNA sequence(5'-3') |
|---|---|---|
| 5' junction PCR | FUT8 5' F | AACTCTGATTTTTGGAATCCCCTTTCTTCAGC |
| FUT8 5' R | TGGGTCTCCCTATAGTGAGTCGTATTAATTTCG | |
| 3' junction PCR | FUT8 3' F | ATGAAGCAGCACGACTTCTTCAAGTCC |
| FUT8 3' R | GCAATGGATGCAAACAGTGGTGTGG | |
| Out-out PCR | FUT8 5' F | AACTCTGATTTTTGGAATCCCCTTTCTTCAGC |
| FUT8 3' R | GCAATGGATGCAAACAGTGGTGTGG |
表3 定点整合验证所需引物
Table 3 Primers for site-specific integration
| 目的 Purpose | 引物名称 Primer name | DNA序列 DNA sequence(5'-3') |
|---|---|---|
| 5' junction PCR | FUT8 5' F | AACTCTGATTTTTGGAATCCCCTTTCTTCAGC |
| FUT8 5' R | TGGGTCTCCCTATAGTGAGTCGTATTAATTTCG | |
| 3' junction PCR | FUT8 3' F | ATGAAGCAGCACGACTTCTTCAAGTCC |
| FUT8 3' R | GCAATGGATGCAAACAGTGGTGTGG | |
| Out-out PCR | FUT8 5' F | AACTCTGATTTTTGGAATCCCCTTTCTTCAGC |
| FUT8 3' R | GCAATGGATGCAAACAGTGGTGTGG |
| 引物名称Primer name | DNA序列DNA sequence(5'-3') |
|---|---|
| M-Bcl2 F | TCACAGAAGGACAAGGTGGATT |
| M-Bcl2 R | AATGCTGACCTGAGCTGGTTT |
| H-Bcl2 F | GAACTGGGGGAGGATTGTGG |
| H-Bcl2 R | CATCCCAGCCTCCGTTATCC |
| GAPDH F | CATGGCCTTCCGTGTTCCTA |
| GAPDH R | TGAAGTCGCAGGAGACAACC |
| FUT8 F | GACCACCCTGACCATTCTAGC |
| FUT8 R | CACGGACTCTTCCTGTAGCTG |
表4 qPCR引物序列
Table 4 Primers for qPCR
| 引物名称Primer name | DNA序列DNA sequence(5'-3') |
|---|---|
| M-Bcl2 F | TCACAGAAGGACAAGGTGGATT |
| M-Bcl2 R | AATGCTGACCTGAGCTGGTTT |
| H-Bcl2 F | GAACTGGGGGAGGATTGTGG |
| H-Bcl2 R | CATCCCAGCCTCCGTTATCC |
| GAPDH F | CATGGCCTTCCGTGTTCCTA |
| GAPDH R | TGAAGTCGCAGGAGACAACC |
| FUT8 F | GACCACCCTGACCATTCTAGC |
| FUT8 R | CACGGACTCTTCCTGTAGCTG |

图2 sgRNA基因编辑效率检测结果 M:DL2000 DNA分子量标准;1:野生型细胞外显子1处PCR产物;2:野生型细胞外显子2处PCR产物;3:转染sgRNA1/Cas9质粒的细胞PCR扩增产物;4:转染sgRNA2/Cas9质粒的细胞PCR扩增产物;5:转染sgRNA3/Cas9质粒的细胞PCR扩增产物
Fig. 2 Results of sgRNA gene editing efficiency M: DL2000 DNA marker. 1: Wild-type cells exon1 PCR amplification products. 2: Wild-type cells exon 2 PCR amplification products. 3: PCR amplification products of sgRNA1/Cas9 transfected cells. 4: Exon1-sgRNA2/Cas9 transfected cells PCR amplification products. 5: PCR amplification products of sgRNA3/Cas9 transfected cells

图4 定点同步双敲细胞分子鉴定结果 A:5'junction PCR电泳结果;B:3'junction PCR电泳结果;C:out-out PCR电泳结果;D:5'/3' junction PCR 扩增产物测序结果。M:DL5000 DNA分子量标准;1:野生型CHO-K1细胞;2:定点同步双敲细胞株
Fig. 4 Molecular identification results of the site-specific synchronous double-knock cell A: Electrophoresis results of 5' junction PCR. B: Electrophoresis results of 5' junction PCR. C: Electrophoresis results of out-out PCR. D: Sequencing results of 5'/3' junction PCR amplification products. M: DL5000 DNA marker. 1: Wildtype CHO-K1 cells. 2: Site-specific synchronous double-knock cell

图5 定点同步双敲细胞基因表达水平 A:内源性Bcl-2基因相对mRNA表达水平;B:Bcl-2基因相对mRNA表达水平;C:FUT8基因相对mRNA表达水平;D:EGFP蛋白表达水平;E:Bcl-2蛋白表达水平;F:FUT8蛋白酶功能。** P<0.01,**** P<0.000 1,n=3,下同
Fig. 5 Gene expressions of the site-specific synchronous double-knock cell A: Relative mRNA expression of endogenous Bcl-2. B: Relative mRNA expression of exogenous Bcl-2. C: Relative mRNA expression of FUT8. D: EGFP protein expression. E: Bcl-2 protein expression. F: FUT8 protease function. ** P<0.01,**** P<0.000 1,n=3. The same below
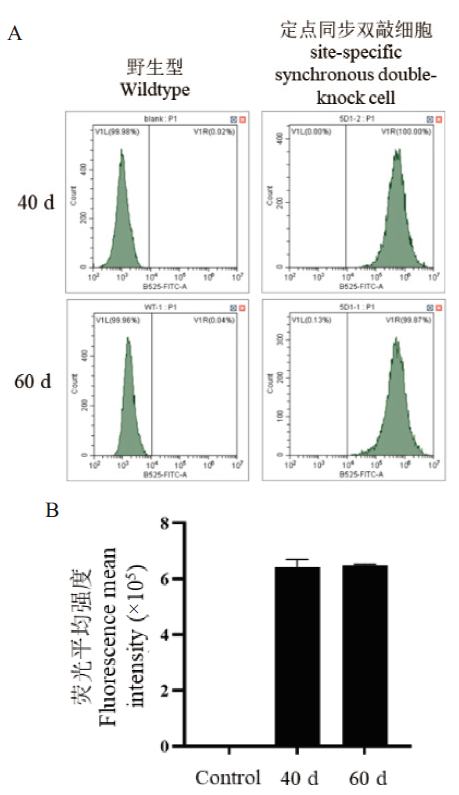
图6 基因表达稳定性结果 A:细胞EGFP蛋白表达比例;B:细胞荧光平均强度
Fig. 6 Stability results of gene expression A: Proportion of EGFP protein expression in cells. B: Fluorescence mean intensity of cells
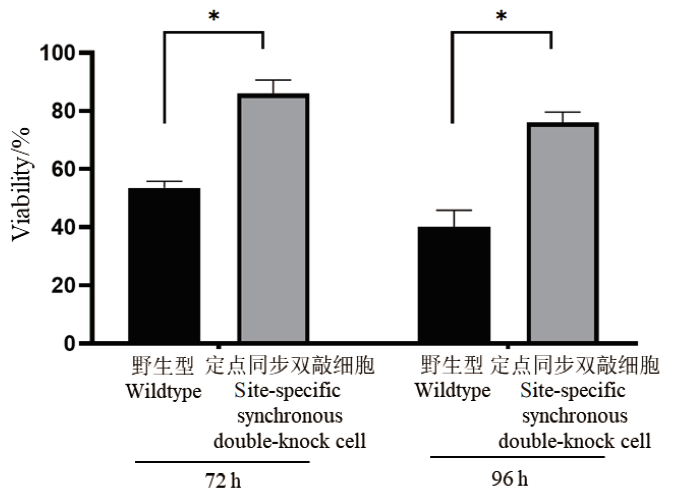
图7 无血清条件下细胞活率 图左:72 h细胞活率;图右:96 h细胞活率。* P<0.05,n=3。下同
Fig. 7 Cell viability under serum-free condition Left part: Cell viability at 72 h. Right part: Cell viability at 96 h. * P<0.05,n=3. The same below

图8 定点同步双敲细胞抗凋亡性能评估 A:坏死细胞比例;B:早期凋亡细胞比例;C:晚期凋亡细胞比例;D:活细胞比例。n=3
Fig. 8 Evaluation on the anti-apoptotic properties of the site-specific synchronous double-knock cell A: Proportion of necrotic cells. B: Proportion of early apoptotic. C: Proportion of late apoptotic. D: Proportion of living cells. n=3
| [1] |
Lin PC, Chan KF, Kiess IA, et al. Attenuated glutamine synthetase as a selection marker in CHO cells to efficiently isolate highly productive stable cells for the production of antibodies and other biologics[J]. mAbs, 2019, 11(5): 965-976.
doi: 10.1080/19420862.2019.1612690 URL |
| [2] |
Hacker DL, Balasubramanian S. Recombinant protein production from stable mammalian cell lines and pools[J]. Curr Opin Struct Biol, 2016, 38: 129-136.
doi: 10.1016/j.sbi.2016.06.005 URL |
| [3] |
Dahodwala H, Lee KH. The fickle CHO: a review of the causes, implications, and potential alleviation of the CHO cell line instability problem[J]. Curr Opin Biotechnol, 2019, 60: 128-137.
doi: 10.1016/j.copbio.2019.01.011 URL |
| [4] |
Ha TK, Kim D, Kim CL, et al. Factors affecting the quality of therapeutic proteins in recombinant Chinese hamster ovary cell culture[J]. Biotechnol Adv, 2022, 54: 107831.
doi: 10.1016/j.biotechadv.2021.107831 URL |
| [5] |
Hartley F, Walker T, Chung V, et al. Mechanisms driving the lactate switch in Chinese hamster ovary cells[J]. Biotechnol Bioeng, 2018, 115(8): 1890-1903.
doi: 10.1002/bit.26603 pmid: 29603726 |
| [6] | Pereira S, Kildegaard HF, Andersen MR. Impact of CHO metabolism on cell growth and protein production: an overview of toxic and inhibiting metabolites and nutrients[J]. Biotechnol J, 2018, 13(3): e1700499. |
| [7] |
Xu X, Nagarajan H, Lewis NE, et al. The genomic sequence of the Chinese hamster ovary(CHO)-K1 cell line[J]. Nat Biotechnol, 2011, 29(8): 735-741.
doi: 10.1038/nbt.1932 URL |
| [8] |
Zhou H, Liu ZG, Sun ZW, et al. Generation of stable cell lines by site-specific integration of transgenes into engineered Chinese hamster ovary strains using an FLP-FRT system[J]. J Biotechnol, 2010, 147(2): 122-129.
doi: 10.1016/j.jbiotec.2010.03.020 pmid: 20371256 |
| [9] |
Fischer S, Handrick R, Otte K. The art of CHO cell engineering: a comprehensive retrospect and future perspectives[J]. Biotechnol Adv, 2015, 33(8): 1878-1896.
doi: 10.1016/j.biotechadv.2015.10.015 pmid: 26523782 |
| [10] |
Lee JS, Grav LM, Lewis NE, et al. CRISPR/Cas9-mediated genome engineering of CHO cell factories: application and perspectives[J]. Biotechnol J, 2015, 10(7): 979-994.
doi: 10.1002/biot.201500082 pmid: 26058577 |
| [11] | Tang DM, Subramanian J, Haley B, et al. Pyruvate kinase muscle-1 expression appears to drive lactogenic behavior in CHO cell lines, triggering lower viability and productivity: a case study[J]. Biotechnol J, 2019, 14(4): e1800332. |
| [12] |
Ha TK, Hansen AH, Kildegaard HF, et al. Knockout of sialidase and pro-apoptotic genes in Chinese hamster ovary cells enables the production of recombinant human erythropoietin in fed-batch cultures[J]. Metab Eng, 2020, 57: 182-192.
doi: S1096-7176(19)30242-3 pmid: 31785386 |
| [13] |
Misaki R, Iwasaki M, Takechi H, et al. Establishment of serum-free adapted Chinese hamster ovary cells with double knockout of GDP-mannose-4, 6-dehydratase and GDP-fucose transporter[J]. Cytotechnology, 2022, 74(1): 163-179.
doi: 10.1007/s10616-021-00501-3 pmid: 35185292 |
| [14] |
Safari F, Farajnia S, Ghasemi Y, et al. Multiplex genome editing in Chinese hamster ovary cell line using all-in-one and HITI CRISPR technology[J]. Adv Pharm Bull, 2021, 11(2): 343-350.
doi: 10.34172/apb.2021.032 pmid: 33880357 |
| [15] |
Tan JGL, Lee YY, Wang TH, et al. Heat shock protein 27 overexpression in CHO cells modulates apoptosis pathways and delays activation of caspases to improve recombinant monoclonal antibody titre in fed-batch bioreactors[J]. Biotechnol J, 2015, 10(5): 790-800.
doi: 10.1002/biot.201400764 pmid: 25740626 |
| [16] |
Lee N, Shin J, Park JH, et al. Targeted gene deletion using DNA-free RNA-guided Cas9 nuclease accelerates adaptation of CHO cells to suspension culture[J]. ACS Synth Biol, 2016, 5(11): 1211-1219.
pmid: 26854539 |
| [17] | Minkenberg B, Wheatley M, Yang YN. CRISPR/Cas9-enabled multiplex genome editing and its application[J]. Prog Mol Biol Transl Sci, 2017, 149: 111-132. |
| [18] |
Wang Q, Betenbaugh MJ. Metabolic engineering of CHO cells to prepare glycoproteins[J]. Emerg Top Life Sci, 2018, 2(3): 433-442.
doi: 10.1042/ETLS20180056 pmid: 33525787 |
| [19] |
Shin SW, Lee JS. Optimized CRISPR/Cas9 strategy for homology-directed multiple targeted integration of transgenes in CHO cells[J]. Biotechnol Bioeng, 2020, 117(6): 1895-1903.
doi: 10.1002/bit.27315 pmid: 32086804 |
| [20] |
Wang WP, Zheng WY, Hu FZ, et al. Enhanced biosynthesis performance of heterologous proteins in CHO-K1 cells using CRISPR-Cas9[J]. ACS Synth Biol, 2018, 7(5): 1259-1268.
doi: 10.1021/acssynbio.7b00375 pmid: 29683658 |
| [21] |
Sakuma T, Yamamoto T. Magic wands of CRISPR-lots of choices for gene knock-in[J]. Cell Biol Toxicol, 2017, 33(6): 501-505.
doi: 10.1007/s10565-017-9409-6 pmid: 28828704 |
| [22] |
Shin SW, Lee JS. CHO cell line development and engineering via site-specific integration: challenges and opportunities[J]. Biotechnol Bioprocess Eng, 2020, 25(5): 633-645.
doi: 10.1007/s12257-020-0093-7 URL |
| [23] |
Yao X, Wang X, Hu XD, et al. Homology-mediated end joining-based targeted integration using CRISPR/Cas9[J]. Cell Res, 2017, 27(6): 801-814.
doi: 10.1038/cr.2017.76 pmid: 28524166 |
| [24] |
Yao X, Zhang ML, Wang X, et al. Tild-CRISPR allows for efficient and precise gene knockin in mouse and human cells[J]. Dev Cell, 2018, 45(4): 526-536.e5.
doi: S1534-5807(18)30327-7 pmid: 29787711 |
| [25] |
Smirnikhina SA, Anuchina AA, Lavrov AV. Ways of improving precise knock-in by genome-editing technologies[J]. Hum Genet, 2019, 138(1): 1-19.
doi: 10.1007/s00439-018-1953-5 pmid: 30390160 |
| [26] |
Lee JS, Grav LM, Pedersen LE, et al. Accelerated homology-directed targeted integration of transgenes in Chinese hamster ovary cells via CRISPR/Cas9 and fluorescent enrichment[J]. Biotechnol Bioeng, 2016, 113(11): 2518-2523.
doi: 10.1002/bit.26002 pmid: 27159230 |
| [1] | 陈小玲, 廖东庆, 黄尚飞, 陈英, 芦志龙, 陈东. 利用CRISPR/Cas9系统改造酿酒酵母的研究进展[J]. 生物技术通报, 2023, 39(8): 148-158. |
| [2] | 杨玉梅, 张坤晓. 应用CRISPR/Cas9技术建立ERK激酶相分离荧光探针定点整合的稳定细胞株[J]. 生物技术通报, 2023, 39(8): 159-164. |
| [3] | 石佳鑫, 刘凯, 朱金洁, 祁显涛, 谢传晓, 刘昌林. 基因编辑技术改良玉米株型增加杂交种产量[J]. 生物技术通报, 2023, 39(8): 62-69. |
| [4] | 施炜涛, 姚春鹏, 魏文康, 王蕾, 房元杰, 仝钰洁, 马晓姣, 蒋文, 张晓爱, 邵伟. 利用CRISPR/Cas9技术构建MDH2敲除细胞株及抗呕吐毒素效应研究[J]. 生物技术通报, 2023, 39(7): 307-315. |
| [5] | 刘晓燕, 祝振亮, 史广宇, 华梓宇, 杨晨, 张涌, 刘军. 乳腺生物反应器的表达优化策略[J]. 生物技术通报, 2023, 39(5): 77-91. |
| [6] | 黄文莉, 李香香, 周炆婷, 罗莎, 姚维嘉, 马杰, 张芬, 沈钰森, 顾宏辉, 王建升, 孙勃. 利用CRISPR/Cas9技术靶向编辑青花菜BoZDS[J]. 生物技术通报, 2023, 39(2): 80-87. |
| [7] | 王兵, 赵会纳, 余婧, 陈杰, 骆梅, 雷波. 利用CRISPR/Cas9系统研究REVOLUTA参与烟草叶芽发育的调控[J]. 生物技术通报, 2023, 39(10): 197-208. |
| [8] | 李双喜, 华进联. 抗猪繁殖与呼吸障碍综合征基因编辑猪研究进展[J]. 生物技术通报, 2023, 39(10): 50-57. |
| [9] | 林蓉, 郑月萍, 徐雪珍, 李丹丹, 郑志富. 拟南芥ACOL8基因在乙烯合成与响应中的功能分析[J]. 生物技术通报, 2023, 39(1): 157-165. |
| [10] | 赖昕彤, 王柯岚, 由雨欣, 谭俊杰. 基于CRISPR/Cas系统的DNA碱基编辑研究进展[J]. 生物技术通报, 2022, 38(6): 1-12. |
| [11] | 刘静静, 刘晓蕊, 李琳, 王盈, 杨海元, 戴一凡. 利用CRISPR/Cas9技术建立OXTR基因敲除猪胎儿成纤维细胞系[J]. 生物技术通报, 2022, 38(6): 272-278. |
| [12] | 张豪, 李哲, 郭凯, 黄艳华, 郝永任. 绿色木霉Tv-1511组蛋白乙酰化酶编码基因TvGCN5的功能分析[J]. 生物技术通报, 2022, 38(5): 136-148. |
| [13] | 陈映丹, 张扬, 夏嫱, 孙虹霞. CRISPR/Cas基因编辑技术及其在微藻研究中的应用[J]. 生物技术通报, 2022, 38(5): 257-268. |
| [14] | Olalekan Amoo, 胡利民, 翟云孤, 范楚川, 周永明. 利用基因编辑技术研究BRANCHED1参与油菜分枝过程的调控[J]. 生物技术通报, 2022, 38(4): 97-105. |
| [15] | 丁亚群, 丁宁, 谢深民, 黄梦娜, 张昱, 张勤, 姜力. Vps28基因敲除小鼠模型的构建及其对泌乳和免疫性状影响的研究[J]. 生物技术通报, 2022, 38(3): 164-172. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||