








生物技术通报 ›› 2021, Vol. 37 ›› Issue (12): 41-49.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0151
收稿日期:2021-02-05
出版日期:2021-12-26
发布日期:2022-01-19
作者简介:徐楠,女,博士,讲师,研究方向:系统代谢工程;E-mail: 基金资助:
XU Nan( ), XU Yu-juan, SUN Pan, ZONG Ren-jie, GUO Min-liang(
), XU Yu-juan, SUN Pan, ZONG Ren-jie, GUO Min-liang( )
)
Received:2021-02-05
Published:2021-12-26
Online:2022-01-19
摘要:
根癌农杆菌(Agrobacterium tumefaciens)能够将自身质粒上的部分DNA片段(简称T-DNA)以T-复合物的形式转运、整合至宿主基因组,而用作高效的植物转基因工具。vbp2是编码VBP蛋白的3个同源基因之一,VBP蛋白通过与T-复合物上的VirD2结合参与T-DNA的转运过程。根据生物信息学预测,vbp2基因的启动子区可能含有多个转录调控元件。通过同源重组的方法,将A. tumefaciens染色体上的vbp2基因替换成编码红色荧光蛋白的rfp基因,使vbp2的启动子控制rfp的表达,因此,可通过测定所表达的RFP的荧光强度来表征vbp2启动子的活性。结果表明:乙酰丁香酮(AS)和鼠李糖(Rha)均能诱导vbp2启动子表达RFP,AS和Rha的最佳诱导浓度分别为150 μmol/L和100 μmol/L。通过截短vbp2启动子DNA片段的方法,证明响应AS和Rha诱导的调控区域分别位于上游的165-260 bp和126-165 bp。
徐楠, 徐宇娟, 孙盼, 宗仁杰, 郭敏亮. 根癌农杆菌vbp2基因启动子转录调控的探析[J]. 生物技术通报, 2021, 37(12): 41-49.
XU Nan, XU Yu-juan, SUN Pan, ZONG Ren-jie, GUO Min-liang. Exploration of the Transcriptional Regulation of Agrobacterium tumefaciens vbp2 Promoter[J]. Biotechnology Bulletin, 2021, 37(12): 41-49.
| 引物 Primer | 序列 Sequence(5' - 3') | 酶切位点 Restriction endonuclease site | 用途 Purpose |
|---|---|---|---|
| Q1-F | TGTGGATCCAACGTCCGCTGCGCGA | BamH I | 扩增vbp2上游526 bp片段 Amplifying upstream 526 bp segment of vbp2 |
| Q1-R | AGCTCCTCGCCCTTGCTCACCATGGGAACGAGGTATCTGACTA | 无None | |
| Rfp-F | CGAAACGAAATAGGTGTACCCATCTGCCAC | 无None | 扩增rfp基因 Amplifying gene rfp |
| Rfp-R | CATGAGCGACGGGAATTCGCCTATCTGTGCCCCAGT | EcoR I | |
| H-F | CTGGGGCACAGATAGCCGCGAGGTGTCAAAGCA | 无None | 扩增vbp2下游1 kb片段 Amplifying downstream 1 kb segment of vbp2 |
| H-R | GAAGCTTATTGCGCTCGGGCCCAGTGGCACGG | Hind III | |
| P-F | CCCAAGCTTGGGAGAGGCGGTTTGCGTATTGGGC | Hind III | 扩增去除pUCA-19质粒的lacZ’启动子 Amplifying lacZ’ promoter with pUCA-19 plasmid removed |
| P-R | CCCAAGCTTACAGGAAACAGCTATGACCATGATT | Hind III | |
| Egfp-F | TAGTCAGATACCTCGTTCCCATGGTGAGCAAGGGCGAGGA | 无None | 扩增gfp序列 Amplifying gfp sequence |
| Egfp-R | CCCAAGCTTTTACTTGTACAGCTCGTCCATGCC | Hind III | |
| Q2-F | CGCGGATCCCGATCCAGTTATAATGAATTTGC | Hind III | 与Q1-R扩增vbp2上游463 bp序列 Amplifying upstream 463 bp sequence of vbp2 with Q1-R |
| Q3-F | CGCGGATCCAAGTAGTAGTAAGGTCGATCG | Hind III | 与Q1-R扩增vbp2上游260 bp序列 Amplifying upstream 260 bp sequence of vbp2 with Q1-R |
| Q4-F | ATCAGGATCCGGCAGCGTGTTTATCAGG | Hind III | 与Q1-R扩增vbp2上游165 bp序列 Amplifying upstream 165 bp sequence of vbp2 with Q1-R |
| Q5-F | CGCGGATCCGACCTACAAAATTCGACTAGCAC | Hind III | 与Q1-R扩增vbp2上游126 bp序列 Amplifying upstream 126 bp sequence of vbp2 with Q1-R |
| Q6-F | CGCGGATCCGGCATTTGATCAACGTGTCG | Hind III | 与Q1-R扩增vbp2上游97 bp序列 Amplifying upstream 97 bp sequence of vbp2 with Q1-R |
| Q7-F | CGCGGATCCATACCACCAGCGCGATTGC | Hind III | 与Q1-R扩增vbp2上游60 bp序列 Amplifying upstream 60 bp sequence of vbp2 with Q1-R |
| Q8-F | CGCGGATCCTAGTCAGATACCTCGTTCCC | Hind III | 与Q1-R扩增vbp2上游20 bp序列 Amplifying upstream 20 bp sequence of vbp2 with Q1-R |
表1 本研究所使用的引物
Table 1 Primers used in this study
| 引物 Primer | 序列 Sequence(5' - 3') | 酶切位点 Restriction endonuclease site | 用途 Purpose |
|---|---|---|---|
| Q1-F | TGTGGATCCAACGTCCGCTGCGCGA | BamH I | 扩增vbp2上游526 bp片段 Amplifying upstream 526 bp segment of vbp2 |
| Q1-R | AGCTCCTCGCCCTTGCTCACCATGGGAACGAGGTATCTGACTA | 无None | |
| Rfp-F | CGAAACGAAATAGGTGTACCCATCTGCCAC | 无None | 扩增rfp基因 Amplifying gene rfp |
| Rfp-R | CATGAGCGACGGGAATTCGCCTATCTGTGCCCCAGT | EcoR I | |
| H-F | CTGGGGCACAGATAGCCGCGAGGTGTCAAAGCA | 无None | 扩增vbp2下游1 kb片段 Amplifying downstream 1 kb segment of vbp2 |
| H-R | GAAGCTTATTGCGCTCGGGCCCAGTGGCACGG | Hind III | |
| P-F | CCCAAGCTTGGGAGAGGCGGTTTGCGTATTGGGC | Hind III | 扩增去除pUCA-19质粒的lacZ’启动子 Amplifying lacZ’ promoter with pUCA-19 plasmid removed |
| P-R | CCCAAGCTTACAGGAAACAGCTATGACCATGATT | Hind III | |
| Egfp-F | TAGTCAGATACCTCGTTCCCATGGTGAGCAAGGGCGAGGA | 无None | 扩增gfp序列 Amplifying gfp sequence |
| Egfp-R | CCCAAGCTTTTACTTGTACAGCTCGTCCATGCC | Hind III | |
| Q2-F | CGCGGATCCCGATCCAGTTATAATGAATTTGC | Hind III | 与Q1-R扩增vbp2上游463 bp序列 Amplifying upstream 463 bp sequence of vbp2 with Q1-R |
| Q3-F | CGCGGATCCAAGTAGTAGTAAGGTCGATCG | Hind III | 与Q1-R扩增vbp2上游260 bp序列 Amplifying upstream 260 bp sequence of vbp2 with Q1-R |
| Q4-F | ATCAGGATCCGGCAGCGTGTTTATCAGG | Hind III | 与Q1-R扩增vbp2上游165 bp序列 Amplifying upstream 165 bp sequence of vbp2 with Q1-R |
| Q5-F | CGCGGATCCGACCTACAAAATTCGACTAGCAC | Hind III | 与Q1-R扩增vbp2上游126 bp序列 Amplifying upstream 126 bp sequence of vbp2 with Q1-R |
| Q6-F | CGCGGATCCGGCATTTGATCAACGTGTCG | Hind III | 与Q1-R扩增vbp2上游97 bp序列 Amplifying upstream 97 bp sequence of vbp2 with Q1-R |
| Q7-F | CGCGGATCCATACCACCAGCGCGATTGC | Hind III | 与Q1-R扩增vbp2上游60 bp序列 Amplifying upstream 60 bp sequence of vbp2 with Q1-R |
| Q8-F | CGCGGATCCTAGTCAGATACCTCGTTCCC | Hind III | 与Q1-R扩增vbp2上游20 bp序列 Amplifying upstream 20 bp sequence of vbp2 with Q1-R |
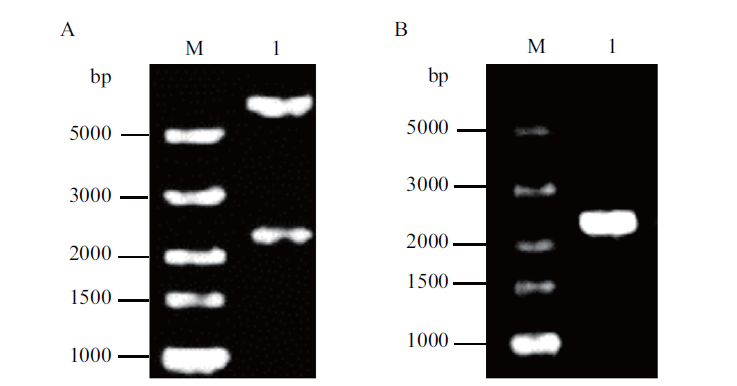
图2 用于进行同源重组构建根癌农杆菌突变体的质粒pEX18Km-rfp的鉴定 A:用构建质粒时所有的限制性内切酶水解质粒所得片段大小;B:用插入片段两端的引物从细菌中扩增得到的PCR产物
Fig.2 Identification of plasmid pEX18Km-rfp used for the construction of A. tumefaciens mutant via homologous recombination A:DNA fragments of the plasmid cleaved by the restriction endonuclease. B:The PCR products amplified from bacteria by primers inserted into the 2 ends of the fragment
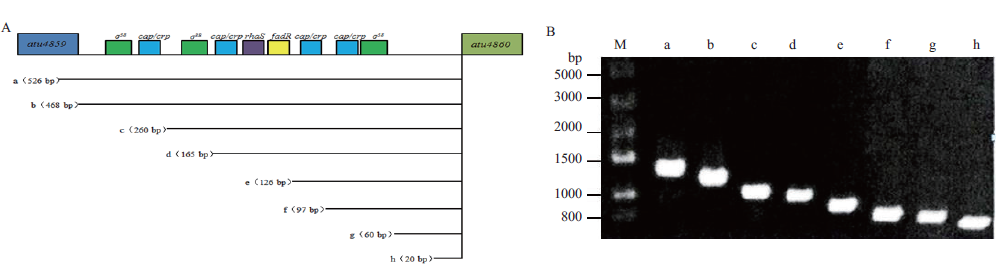
图5 受不同长度的vbp2启动子片段控制的egfp表达质粒的构建和鉴定。 A:被截短的vbp2启动子片段的长度、位置及可能存在的调控元件;B:用PCR鉴定这些携带受不同长度的vbp2启动子片段控制的egfp表达质粒的根癌农杆菌菌株
Fig.5 Construction and identification of the egfp expression plasmids controlled by different lengths of vbp2 promotor A:The length,region and possible regulatory elements of the truncated vbp2 promotor fragment. B:PCR-identified A. tumefaciens strains that carry plasmids expressing egfp under the control of different lengths of vbp2 promotor
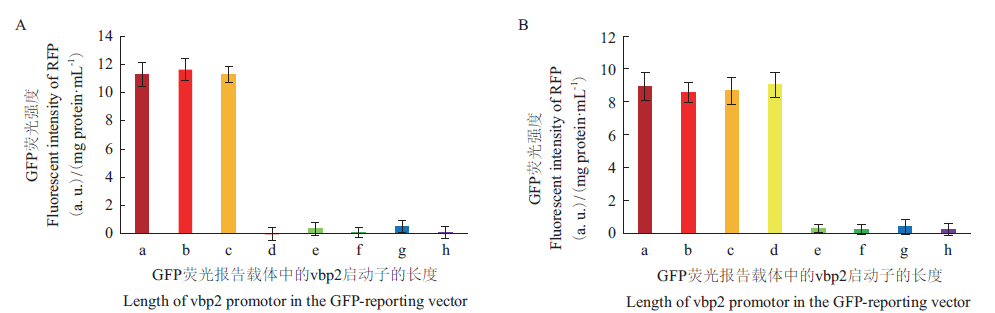
图6 分别以150 μmol/L AS(A)和100 μmol/L Rha(B)诱导不同长度vbp2启动子表达GFP的情况
Fig. 6 GFP expression promoted by different lengths of vbp2 promotor under the induction of 150 μmol/L AS(A)or 100 μmol/L Rha(B)
| [1] |
Guo ML, Ye JY, Gao DW, et al. Agrobacterium-mediated horizontal gene transfer:Mechanism, biotechnological application, potential risk and forestalling strategy[J]. Biotechnol Adv, 2019, 37(1):259-270.
doi: 10.1016/j.biotechadv.2018.12.008 URL |
| [2] |
Matveeva TV, Lutova LA. Horizontal gene transfer from Agrobacterium to plants[J]. Front Plant Sci, 2014, 5:326.
doi: 10.3389/fpls.2014.00326 pmid: 25157257 |
| [3] | Guo ML, Bian XW, Wu X, et al. Agrobacterium-mediated genetic transformation:history and progress[M]// Genetic Transformation. InTech, 2011:3-28. |
| [4] | 叶文兴, 孔令琪. 影响根癌农杆菌转化效率的因素综述[J]. 中国草地学报, 2019, 41(3):142-148. |
| Ye WX, Kong LQ. Advanced in factors influencing Agrobacterium tumefaciens-mediates transformation efficiency[J]. Chin J Grassland, 2019, 41(3):142-148. | |
| [5] | 王根平, 杜文明, 夏兰琴. 植物安全转基因技术研究现状与展望[J]. 中国农业科学, 2014, 47(5):823-843. |
| Wang GP, Du WM, Xia LQ. Current status of transgenic technologies for safety consideration in plants and future perspectives[J]. Sci Agric Sin, 2014, 47(5):823-843. | |
| [6] | 赵佩, 王轲, 张伟, 等. 参与农杆菌侵染及T-DNA转运过程植物蛋白的研究进展和思考[J]. 中国农业科学, 2014, 47(13):2504-2518. |
| Zhao P, Wang K, Zhang W, et al. Review and inspiration of plant proteins involved in the transformation processing of T-DNA initiated by Agrobacterium[J]. Sci Agric Sin, 2014, 47(13):2504-2518. | |
| [7] |
Ankenbauer RG, Nester EW. Sugar-mediated induction of Agrobacterium tumefaciens virulence genes:structural specificity and activities of monosaccharides[J]. J Bacteriol, 1990, 172(11):6442-6446.
pmid: 2121715 |
| [8] |
Subramoni S, Nathoo N, Klimov E, et al. Agrobacterium tumefaciens responses to plant-derived signaling molecules[J]. Front Plant Sci, 2014, 5:322.
doi: 10.3389/fpls.2014.00322 pmid: 25071805 |
| [9] |
Stachel SE, Messens E, van Montagu M, et al. Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens[J]. Nature, 1985, 318(6047):624-629.
doi: 10.1038/318624a0 URL |
| [10] | 郭敏亮, 高佃坤, 金英善. 农杆菌T-复合物的形成与转运研究进展[J]. 生物化学与生物物理进展, 2009, 36(11):1408-1414. |
|
Guo ML, Gao DK, Jin YS. Progress in the formation and transfer of Agrobacterium T-complex[J]. Prog Biochem Biophys, 2009, 36(11):1408-1414.
doi: 10.3724/SP.J.1206.2009.00195 URL |
|
| [11] |
Binns AN, Thomashow MF. Cell biology of Agrobacterium infection and transformation of plants[J]. Annu Rev Microbiol, 1988, 42(1):575-606.
doi: 10.1146/micro.1988.42.issue-1 URL |
| [12] | Lacroix B, Citovsky V. Transfer of DNA from bacteria to eukaryotes[J]. mBio, 2016, 7(4):e00863-e00816. |
| [13] |
Guo ML, Hou QM, Hew CL, et al. Agrobacterium VirD2-binding protein is involved in tumorigenesis and redundantly encoded in conjugative transfer gene clusters[J]. Mol Plant Microbe Interact, 2007, 20(10):1201-1212.
doi: 10.1094/MPMI-20-10-1201 URL |
| [14] |
Guo ML, Jin SG, Sun DY, et al. Recruitment of conjugative DNA transfer substrate to Agrobacterium type IV secretion apparatus[J]. PNAS, 2007, 104(50):20019-20024.
doi: 10.1073/pnas.0701738104 URL |
| [15] | 郭敏亮. 农杆菌T-复合物招募蛋白的发现和生物学功能鉴定[J]. 江西农业大学学报, 2010, 32(5):992-996. |
| Guo ML. Discovery and biological function identification of Agrobacterium T-complex recruiting protein[J]. Acta Agric Univ Jiangxiensis, 2010, 32(5):992-996. | |
| [16] | Yang J, Wu MX, et al. Expression of Agrobacterium homolog genes encoding T-complex recruiting protein under virulence induction conditions[J]. Front Microbiol, 2015, 6:1379. |
| [17] |
Guo ML, Zhu Q, Gao DK. Development and optimization of method for generating unmarked A. tumefaciens mutants[J]. Prog Biochem Biophys, 2009, 36(5):556-565.
doi: 10.3724/SP.J.1206.2008.00618 URL |
| [18] | 郭敏亮, 姜涌明. 考马斯亮蓝显色液组分对蛋白质测定的影响[J]. 生物化学与生物物理进展, 1996, 23(6):558-561. |
| Guo ML, Jiang YM. Effect of ingredients of coomassie brilliant blue color-developing reagent on protein assay[J]. Prog Biochem Biophys, 1996, 23(6):558-561. | |
| [19] | 樊晋宇, 崔宗强, 张先恩. 红色荧光蛋白的光谱多样性及体外分子进化[J]. 生物化学与生物物理进展, 2008, 35(10):1112-1120. |
| Fan JY, Cui ZQ, Zhang XE. Optical spectra diversity and in vitro molecular evolution of red fluorescent proteins[J]. Prog Biochem Biophys, 2008, 35(10):1112-1120. | |
| [20] |
Merzlyak EM, Goedhart J, Shcherbo D, et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime[J]. Nat Methods, 2007, 4(7):555-557.
doi: 10.1038/nmeth1062 URL |
| [21] |
王飞, 杨海涛, 王泽方. 红色荧光蛋白的研究进展[J]. 生物技术通报, 2017, 33(9):32-47.
doi: 10.13560/j.cnki.biotech.bull.1985.2017-0401 |
| Wang F, Yang HT, Wang ZF. Research progress on red fluorescent protein[J]. Biotechnol Bull, 2017, 33(9):32-47. | |
| [22] |
Zimmer M. Green fluorescent protein(GFP):applications, structure, and related photophysical behavior[J]. Chem Rev, 2002, 102(3):759-781.
pmid: 11890756 |
| [23] |
Miesenböck G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins[J]. Nature, 1998, 394(6689):192-195.
doi: 10.1038/28190 URL |
| [24] |
Dong T, Schellhorn HE. Role of RpoS in virulence of pathogens[J]. Infect Immun, 2010, 78(3):887-897.
doi: 10.1128/IAI.00882-09 pmid: 19948835 |
| [25] |
Caimano MJ, Groshong AM, Belperron A, et al. The RpoS gatekeeper in Borrelia burgdorferi:an invariant regulatory scheme that promotes spirochete persistence in reservoir hosts and niche diversity[J]. Front Microbiol, 2019, 10:1923.
doi: 10.3389/fmicb.2019.01923 URL |
| [26] |
Huang L, Guo L, Xu X, et al. The role of rpoS in the regulation of Vibrio alginolyticus virulence and the response to diverse stresses[J]. J Fish Dis, 2019, 42(5):703-712.
doi: 10.1111/jfd.2019.42.issue-5 URL |
| [27] | 陈翠娜, 冯德玉, 张新, 等. 乙酰丁香酮及共培养基pH对棉花遗传转化的影响[J]. 上海交通大学学报:农业科学版, 2013, 31(1):18-22. |
| Chen CN, Feng DY, Zhang X, et al. Effect of acetosyringone and media pH on transformation of cotton[J]. J Shanghai Jiao Tong Univ:Agric Sci, 2013, 31(1):18-22. | |
| [28] |
Guo ML, Huang ZW, Yang J. Is there any crosstalk between the chemotaxis and virulence induction signaling in Agrobacterium tumefaciens?[J]. Biotechnol Adv, 2017, 35(4):505-511.
doi: 10.1016/j.biotechadv.2017.03.008 URL |
| [29] | Wise AA, Binns AN. The receiver of the Agrobacterium tumefaciens VirA histidine kinase forms a stable interaction with VirG to activate virulence gene expression[J]. Front Microbiol, 2015, 6:1546. |
| [30] |
Tobin JF, Schleif RF. Purification and properties of RhaR, the positive regulator of the L-rhamnose operons of Escherichia coli[J]. J Mol Biol, 1990, 211(1):75-89.
pmid: 2405166 |
| [31] |
Wickstrum JR, Skredenske JM, Balasubramaniam V, et al. The AraC/XylS family activator RhaS negatively autoregulates rhaSR expression by preventing cyclic AMP receptor protein activation[J]. J Bacteriol, 2010, 192(1):225-232.
doi: 10.1128/JB.00829-08 pmid: 19854903 |
| [32] |
Kolin A, Balasubramaniam V, Skredenske JM, et al. Differences in the mechanism of the allosteric l-rhamnose responses of the AraC/XylS family transcription activators RhaS and RhaR[J]. Mol Microbiol, 2008, 68(2):448-461.
doi: 10.1111/j.1365-2958.2008.06164.x pmid: 18366439 |
| [1] | 李英, 岳祥华. DNA甲基化在解析毛竹自然变异中的应用[J]. 生物技术通报, 2023, 39(7): 48-55. |
| [2] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [3] | 陈强, 邹明康, 宋家敏, 张冲, 吴隆坤. 甜瓜LBD基因家族的鉴定和果实发育进程中的表达分析[J]. 生物技术通报, 2023, 39(3): 176-183. |
| [4] | 许睿, 祝英方. 中介体复合物在植物非生物胁迫应答中的功能[J]. 生物技术通报, 2023, 39(11): 54-60. |
| [5] | 张婵, 吴友根, 于靖, 杨东梅, 姚广龙, 杨华庚, 张军锋, 陈萍. 光与茉莉酸信号介导的萜类化合物合成分子机制[J]. 生物技术通报, 2022, 38(8): 32-40. |
| [6] | 周承哲, 常笑君, 朱晨, 程春振, 陈裕坤, 赖钟雄, 林玉玲, 郭玉琼. 茶树原位转化方法的建立[J]. 生物技术通报, 2022, 38(2): 263-268. |
| [7] | 陈臣, 黄芝阳, 于海燕, 袁海彬, 田怀香. 原核生物转录调控研究技术及进展[J]. 生物技术通报, 2022, 38(10): 54-65. |
| [8] | 张小妮, 翁伊纯, 范奕浩, 王晓娟, 赵佳宇, 张云龙. Mito-OS-Timer:一种靶向监测线粒体氧化应激的荧光秒表[J]. 生物技术通报, 2022, 38(10): 97-105. |
| [9] | 杨威, 伍茜, 程建国, 罗燕, 王印, 杨泽晓, 姚学萍. 林麝干扰素α基因克隆、表达及转录调控分析[J]. 生物技术通报, 2022, 38(1): 194-204. |
| [10] | 王欢禹, 常昊宛, 章崇祺, 金卫林, 魏芳. 五种检测嵌合抗原受体表达方法的比较[J]. 生物技术通报, 2021, 37(12): 265-273. |
| [11] | 马军, 徐通达. 植物非经典生长素信号转导通路解析[J]. 生物技术通报, 2020, 36(7): 15-22. |
| [12] | 刘文浩, 王瑞丰, 刘琬琳, 许杰. 不同调控元件及组合对烟草外源蛋白瞬时表达的效果分析[J]. 生物技术通报, 2020, 36(7): 62-71. |
| [13] | 王彩霞, 杜方原, 林祥梅, GrzegorzWozniakowski, 王勤, 冯春燕, 吴绍强. 稳定表达非洲猪瘟病毒P54蛋白的Vero细胞系的建立[J]. 生物技术通报, 2020, 36(5): 139-144. |
| [14] | 陈永灿, 张建志, 司同. 酿酒酵母中基于CRISPR/dCás9的基因转录调控工具的开发与应用[J]. 生物技术通报, 2020, 36(4): 1-12. |
| [15] | 李晓佩, 王思宁, 史晶晶, 高志民. 植物表皮蜡质合成及调控因子WIN/SHN的研究进展[J]. 生物技术通报, 2020, 36(12): 129-136. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||